- Title
-
Knockout of Nur77 Leads to Amino Acid, Lipid, and Glucose Metabolism Disorders in Zebrafish
- Authors
- Xu, Y., Tian, J., Kang, Q., Yuan, H., Liu, C., Li, Z., Liu, J., Li, M.
- Source
- Full text @ Front Endocrinol (Lausanne)
|
Knockout of PHENOTYPE:
|
|
RNA-seq (RNA sequencing) analysis of |
|
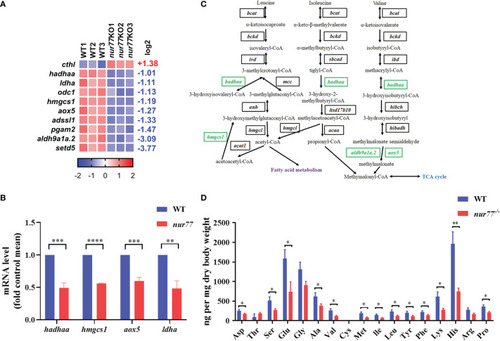
Nur77 regulates amino acid metabolism in zebrafish larvae. EXPRESSION / LABELING:
PHENOTYPE:
|
|
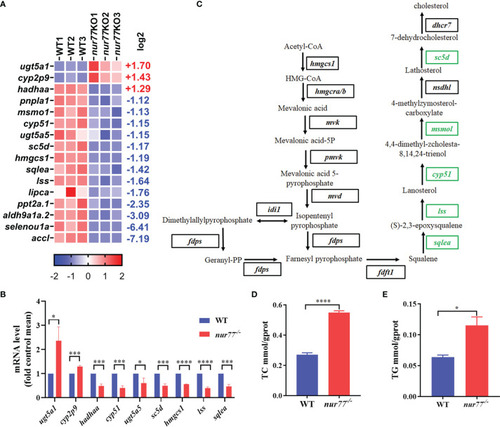
Nur77 regulates lipid metabolism in zebrafish larvae. |
|
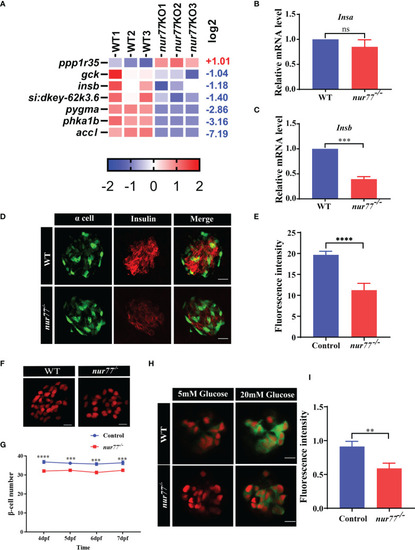
Nur77 regulates carbohydrate metabolism in zebrafish larvae. |
|
Nur77 regulates insulin secretion and β-cell number in zebrafish larvae. |
|
Targeted expression of Nur77 in β cells restored β-cell number and insulin content in zebrafish. |