- Title
-
Vertebrate-specific glutaredoxin is essential for brain development
- Authors
- Bräutigam, L., Schütte, L.D., Godoy, J.R., Prozorovski, T., Gellert, M., Hauptmann, G., Holmgren, A., Lillig, C.H., and Berndt, C.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
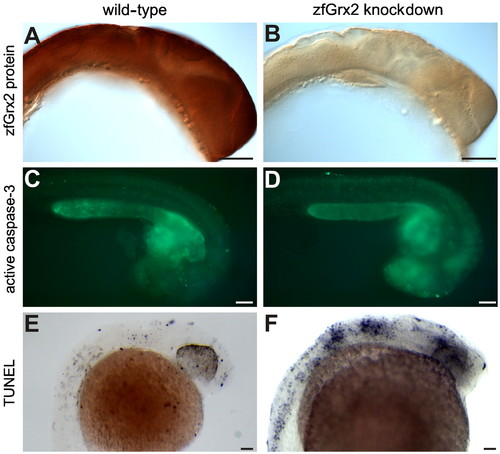
Knockdown of zfGrx2 induced apoptosis in central nervous system. Injection of a morpholino blocking zfGrx2 translation reduced the levels of the corresponding protein as visualized by whole mount immunohistochemistry (A and B). Loss of zfGrx2 induced apoptosis in the central nervous system as demonstrated by in vivo caspase 3 measurements (C and D) and TUNEL staining (E and F). Scale bar, 50 µm. |
|
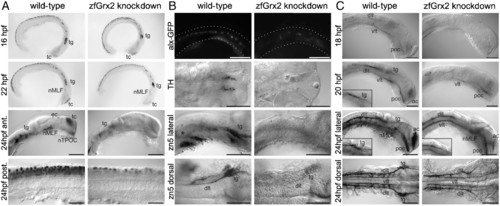
Knockdown of zfGrx2 suppressed formation of a neuronal network. Morpholino-induced knockdown of zfGrx2 led to loss of early HuC/D positive neurons between 22 and 24 hpf (A). As demonstrated by different techniques, all tested neuronal subgroups were affected at 24 hpf: glutamatergic excitatory interneurons, dopaminergic neurons, and secondary motor neurons (B). Embryos lacking zfGrx2 were not able to develop an axonal scaffold (C). Ac, anterior commissure; dlt, dorsal longitudinal tract; nMLF, nucleus of the medial longitudinal fascicle; poc, posterior commissure; tg, trigeminal ganglion; vlt, ventral longitudinal tract. Scale bar, 50 µm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Outgrowth of neurites depends on the presence of Grx2. (A) Length and number of branching points of axons in embryos with silenced zfGrx2 expression were dramatically reduced 24 hpf, as demonstrated by immunohistochemistry using antiacetylated tubulin antibodies (asterisks mark cell bodies). (B) Quantitative data obtained from A. White bars, wild-type; black bars, zfGrx2 knockdown (mean ± SEM, n = 25, two-tailed Student’s t test, *** p < 3 × 10-7). (C) Three-dimensional reconstruction of the cytoskeleton (red, DNA; blue, tubulin; green, actin; single confocal layers as Insets; blue, DNA; yellow, actin) of SH-SY5Y cells grown for 8 d +/- RA. (D) Quantification of axon lengths (RA, n = 247; +RA, n = 156) and branching points (RA, n = 106; +RA, n = 213) of three independent experiments corresponding to C; white bars, control cells; black bars, hGrx2c+ cells (mean ± SEM, two-tailed Student’s t test, ***p < 0.008). PHENOTYPE:
|
|
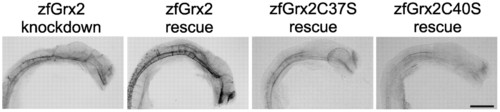
Formation of the axonal network depends on oxidoreductase activity of zfGrx2. Impaired formation of an axonal network (antiacetylated tubulin antibodies) could be rescued by injection of 20 pg/embryo capped zfGrx2 mRNA simultaneously to morpholino injection. Injection of mRNA coding for active site mutants of zfGrx2 (zfGrx2C37S, zfGrx2C40S) did not rescue. Scale bar, 50 µm. PHENOTYPE:
|
|
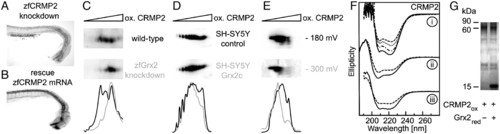
Grx2 acts via CRMP2. Knockdown of zfCRMP2 by morpholino injection impaired formation of axon scaffold (A). Impaired formation of axon scaffold by knockdown of zfGrx2 was rescued by simultaneous injection of 20 pg/embryo capped zfCRMP2 mRNA (B). Densitometric analyses of the resulting pattern of CRMP2-specific spots after separation by 2D redox blots demonstrated Grx2-dependent changes in the thiol redox state of CRMP2 (C-E). At 24 hpf zfCRMP2 was more oxidized in embryos lacking zfGrx2 compared to wild-type (C), whereas in SH-SY5Y cells overexpressing hGrx2c the redox state of CRMP2 was more reduced compared to control cells (D). The redox state of recombinant zfCRMP2 was more reduced after incubation with recombinant zfGrx2 for 1 h under anaerobic conditions in a GSH/GSSG buffer adjusted to -300 mV compared to a buffer with a potential of -180 mV (E). Thiol redox modifications changed secondary structure of zfCRMP2 (F). (F, I) Ellipticity of reduced recombinant zfCRMP2 (solid line) and after incubation with H2O2 for 1 min (dashed/dotted line), 15 min (dotted line), and 30 min (dashed line). (F, ii) Spectra of oxidized zfCRMP2 before (dashed line) and after incubation with GSH/zfGrx2 (solid line). (F, iii) Spectra of oxidized zfCRMP2 before (dashed line) and after incubation with reduced zfGrx2 at a ratio of 161.2 (solid line). (G) A Coomassie blue-stained PAGE comparing quarternary structure of oxidized zfCRMP (60 kDa) before and after incubation with reduced zfGrx2 (16 kDa) at a ratio of 161.2. PHENOTYPE:
|
|
Transcription pattern of zebrafish (zf) glutaredoxin (Grx) 2. In situ hybridization against zfGrx2 at different stages [(A) four cell stage, (B) five somites, and (C) 24 hours post fertilization] showed ubiquitous distribution of zfGrx2 transcripts throughout embryonic development. |
|
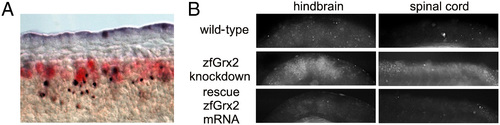
Neuronal apoptosis in embryos is specific for the lack of zebrafish (zf) glutaredoxin (Grx) 2. Costaining of neurons (anti-HuC/D antibody, red) and apoptotic cell death (TUNEL assay, blue) at 24 hours post fertilization (hpf) in embryos with blocked zfGrx2 expression. (A) Apoptosis at 24 hpf in the CNS of embryos lacking zfGrx2 detected by active caspase-3 measurements was rescued by injection of 20 pgembryo capped zfGrx2 mRNA simultaneously to morpholino injection. PHENOTYPE:
|
|
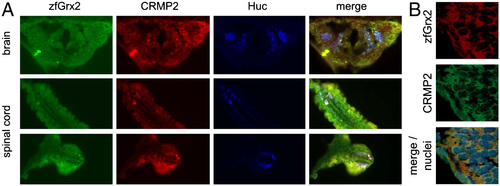
Colocalization of zebrafish (zf) glutaredoxin (Grx) 2 and zebrafish collapsin response mediator protein 2 (CRMP2) in the central nervous system. Immunohistochemistry of 24 hours post fertilization (hpf) zebrafish embryos using anti-HuC/D (early pan neurons, blue), anti-zfGrx2 (green), and anti-CRMP2 antibodies (red) demonstrated colocalization of zfGrx2 and zfCRMP2 especially in neurons (A). B indicated colocalization of zfGrx2 (red) and zfCRMP2 (green) in the cytosol. Nuclei were counterstained with Hoechst 33342 (blue). EXPRESSION / LABELING:
|

Unillustrated author statements PHENOTYPE:
|