- Title
-
In vivo measurement of an Apelin gradient with a genetically encoded APLNR conformation biosensor
- Authors
- Herdt, L., Schihada, H., Kurz, M., Ernst, S., Eberlein, J., Kolb, P., Krasel, C., Bünemann, M., Helker, C.S.M.
- Source
- Full text @ Nat. Commun.
|
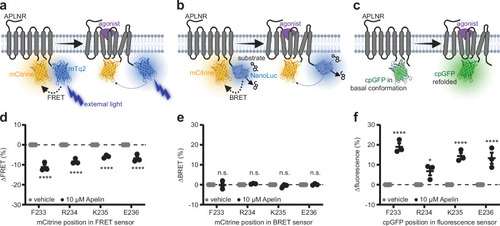
APLNR conformational biosensor design and validation. Schematic illustration of the APLNR conformational biosensors design based on FRET ( |
|
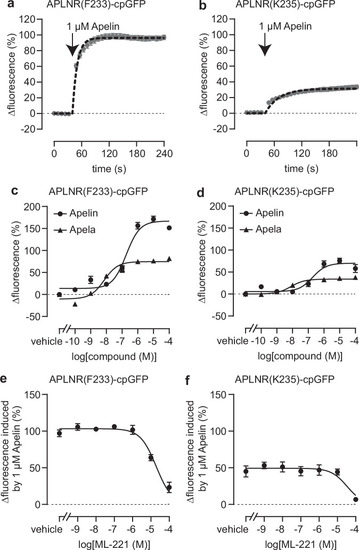
Pharmacological characterization of the APLNR-cpGFP biosensors. Stimulation with Apelin and Apela (arrow indicates timepoint of ligand addition) led to a significant increase of APLNR(F233)-cpGFP ( |
|
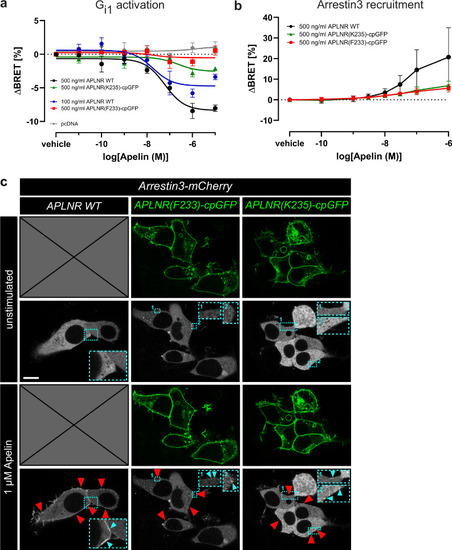
APLNR-cpGFP biosensors possess signaling ability. Gi1 protein dissociation ( |
|
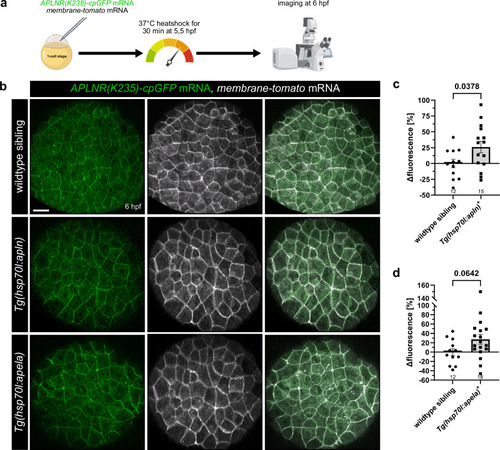
Ubiquitous Apelin and Apela overexpression activate APLNR(K235)-cpGFP biosensor in vivo. |
|
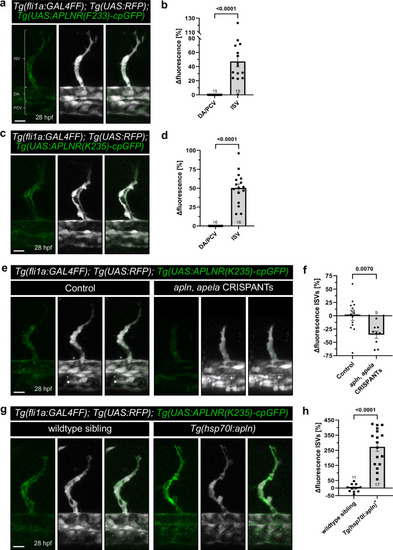
APLNR-cpGFP biosensors visualize endogenous Aplnr activity in vivo. |
|
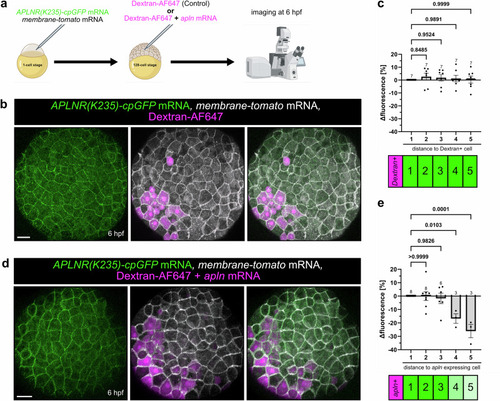
Measuring an Apelin ligand gradient in vivo. |
|
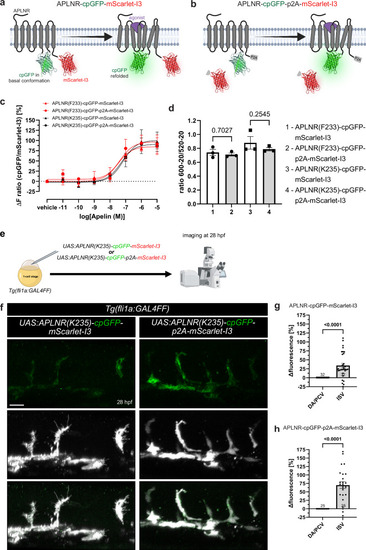
Development and in vivo application of ratiometric APLNR-cpGFP-mScarlet-I3 biosensors. |