- Title
-
Melanoma innervation, noradrenaline and cancer progression in zebrafish xenograft model
- Authors
- Lorenzini, F., Marines, J., Le Friec, J., Do Khoa, N., Nieto, M.A., Sanchez-Laorden, B., Mione, M.C., Fontenille, L., Kissa, K.
- Source
- Full text @ Cell Death Discov
|
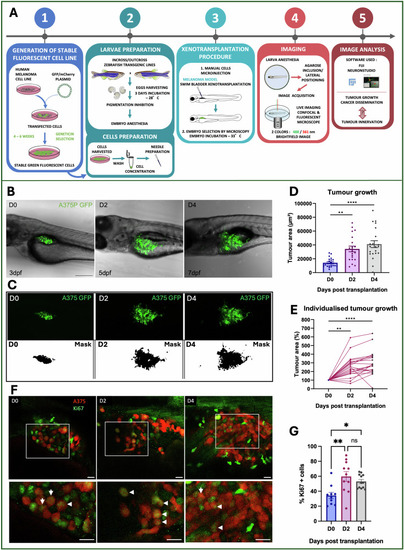
Evaluation of in vivo melanoma growth. |
|
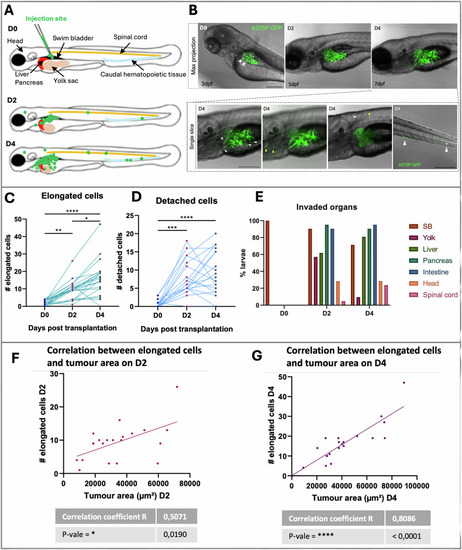
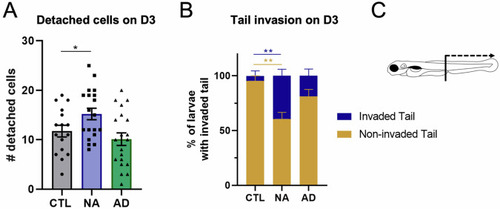
In vivo invasiveness and dissemination of A375P cells after xenografting in larval zebrafish. |
|
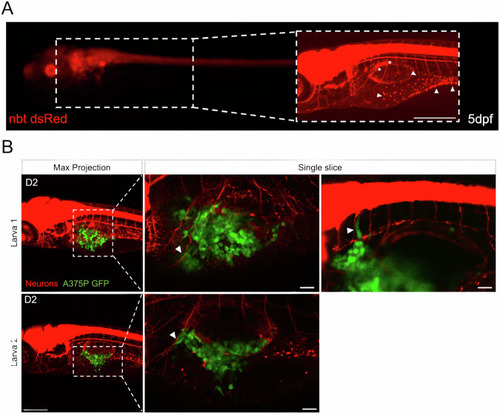
Interactions between PNS and transplanted melanoma cells in larval zebrafish. |
|
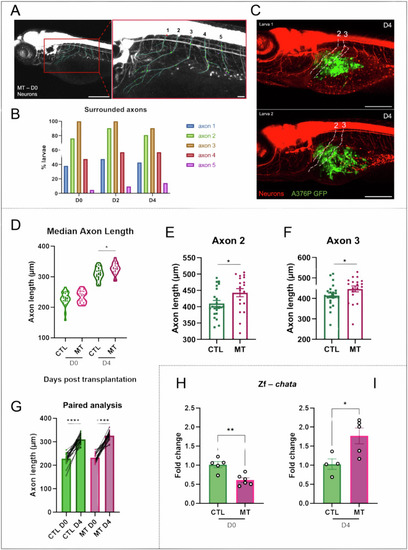
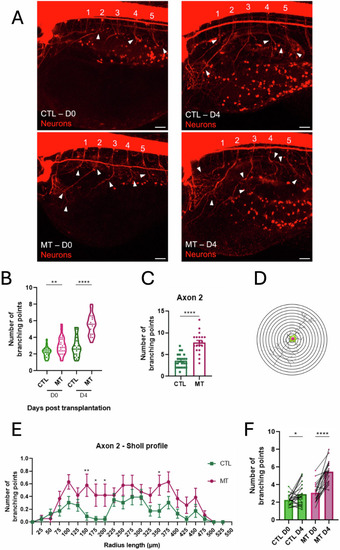
Axonogenesis in zebrafish xenograft melanoma model. |
|
Dendritogenesis in zebrafish xenograft melanoma model. |
|
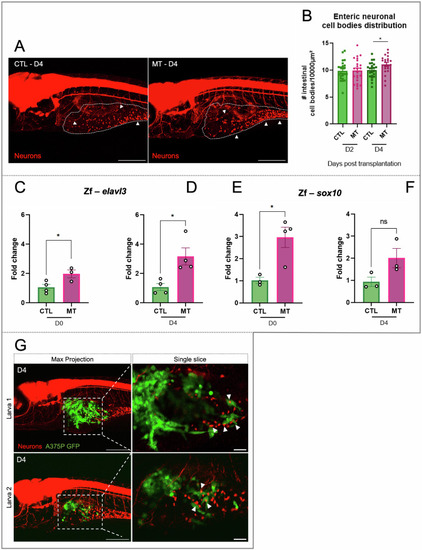
Neurogenesis in zebrafish xenograft melanoma model. |
|
Effect of |