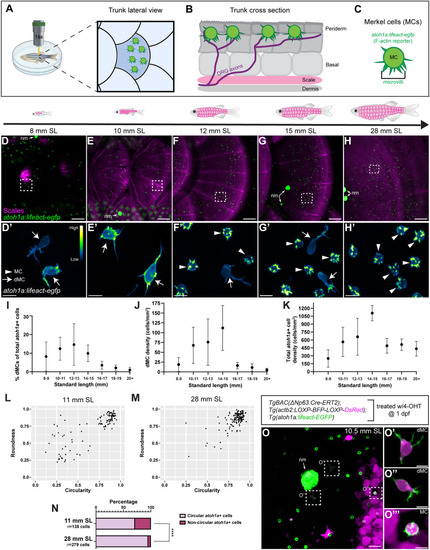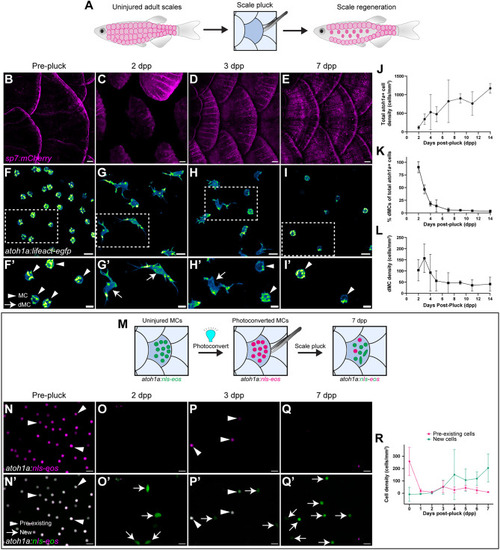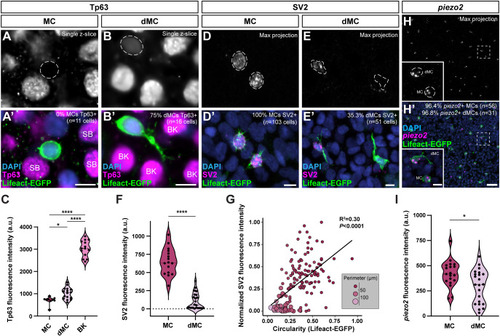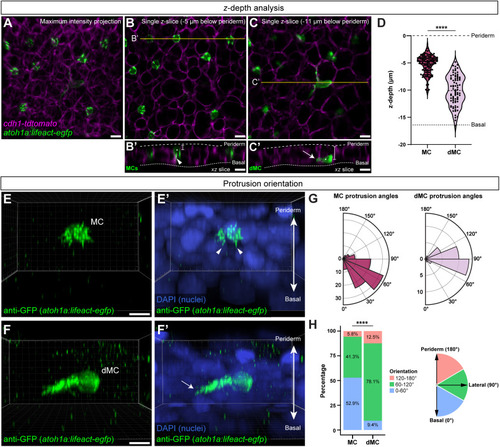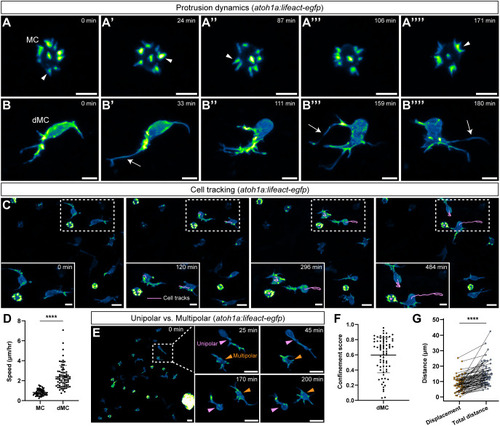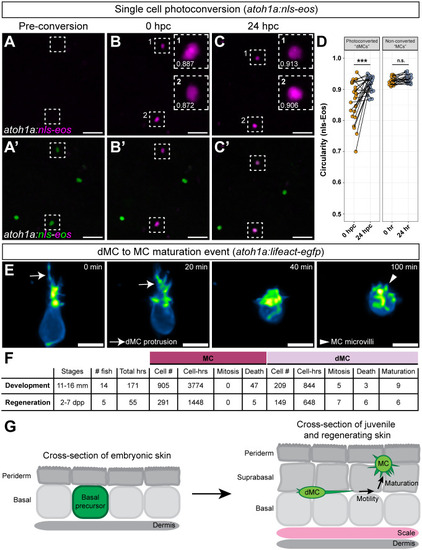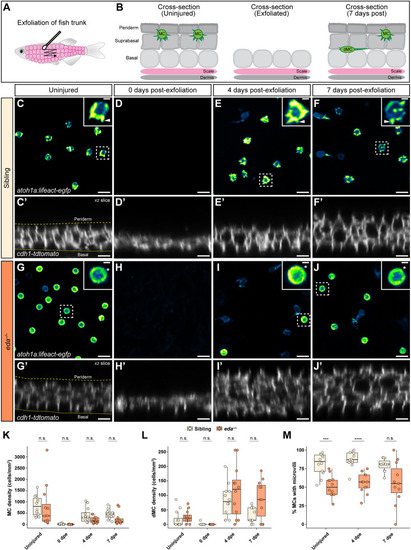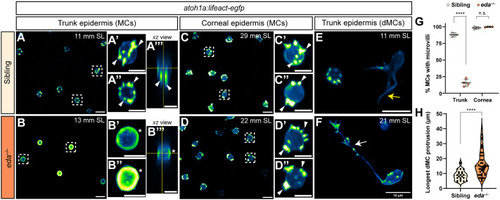
Genetic loss of Eda results in altered dMC and MC morphologies in trunk skin. (A-B‴) Representative confocal images of MCs within the trunk epidermis of the indicated genotypes. Dashed boxes indicate cells magnified in A′,A″,B′,B″. Arrowheads indicate microvilli. Asterisks indicate intense Lifeact-EGFP signal forming a smooth ring-like cortical structure evident in cross-section in the eda−/− mutant epidermis. (A‴,B‴) xz views of the cells shown in A′ and B′. (C-D″) Representative images of MCs within corneal epidermis of animals of the indicated genotypes. Dashed boxes indicate cells magnified in C′,C″,D′,D″. Arrowheads indicate microvilli. (E,F) Representative images of dMCs within the trunk epidermis of the indicated genotypes. Arrows indicate the longest protrusion on a dMC in eda sibling (E) or eda−/− mutant (F) epidermis. (G) Quantification of the percentage of MCs with discernable microvilli in trunk or corneal epidermis of the indicated genotypes. Each dot indicates an animal (9-20 mm SL) where a collection of images was analyzed (sibling trunk, n=386 cells from three fish; eda−/− trunk, n=235 cells from four fish; sibling cornea, n=169 cells from four fish; eda−/− cornea, n=332 cells from four fish). Fisher's exact test shows a significant difference between genotypes in the trunk but not cornea (****P<0.0001; n.s., P=0.3413). (H) Violin plots of the longest dMC Lifeact-EGFP+ protrusion within the trunk epidermis of juveniles of the indicated genotypes. Each dot represents a cell (siblings, n=22 dMCs from six fish; eda−/−, n=47 dMCs from four fish). A non-parametric Mann–Whitney test (****P<0.0001) was used to compare between cell types. Scales bars: 10 µm in A-F; 5 µm in A′-A‴,B′-B‴,C′-C″,D′-D″.
|