- Title
-
KDM4C works in concert with GATA1 to regulate heme metabolism in head and neck squamous cell carcinoma
- Authors
- Wu, M.J., Yang, S.M., Fang, W.K., Chen, T.J., Wu, C.Y., Hsu, Y.J., Shen, C.E., Cheng, Y.C., Hsieh, W.C., Yuh, C.H., Yang, M.H., Kung, H.J., Wang, W.C.
- Source
- Full text @ Cell. Mol. Life Sci.
|
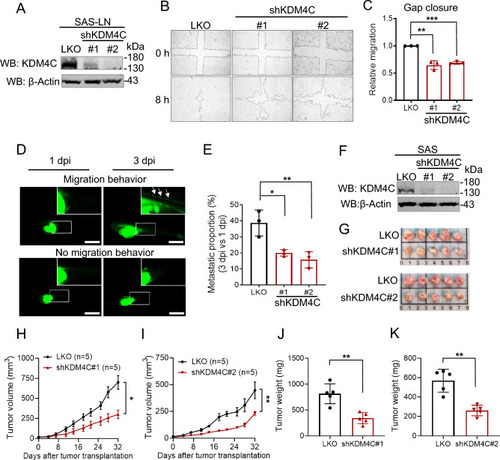
KDM4C promotes cell migration and is crucial for tumor growth in HNSCC. |
|
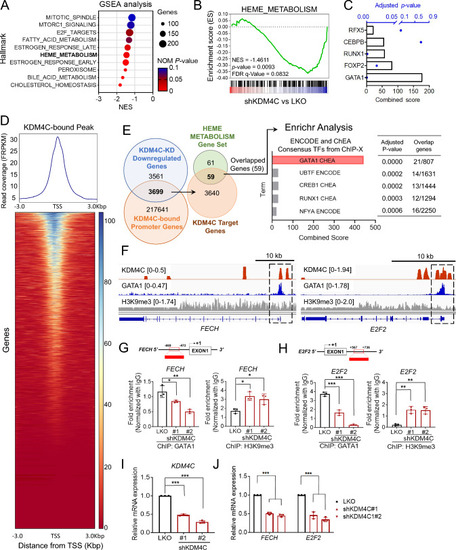
Heme-metabolism genes are regulated by KDM4C and GATA1. |
|
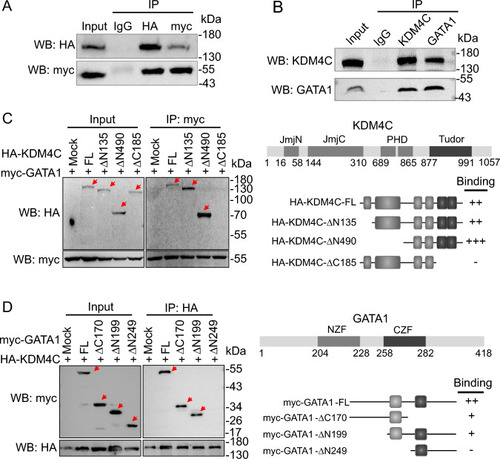
The interaction of KDM4C and GATA1. |
|
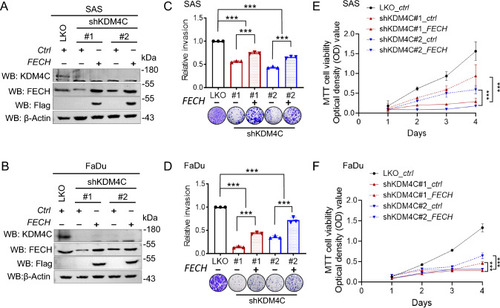
Effect of FECH overexpression on invasion and proliferation in KDM4C-knockdown SAS and FaDu cells. |
|
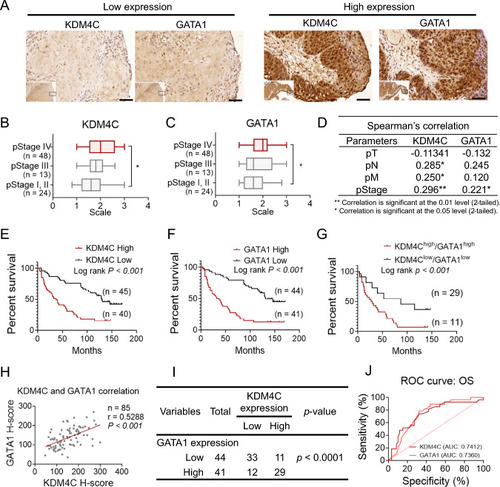
Correlation and clinical impact of KDM4C and GATA1 expression in HNSCC. |
|
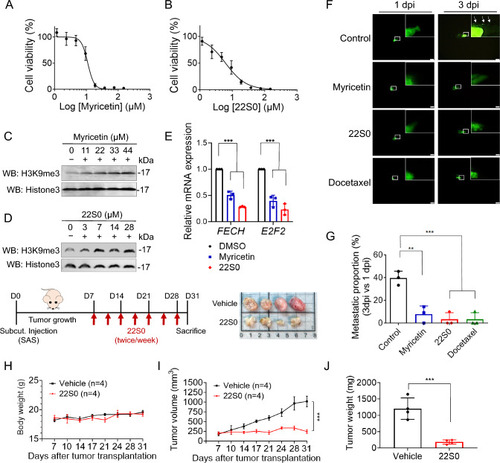
Effects of KDM4 inhibitors on heme metabolism and tumor growth in HNSCC. |
|
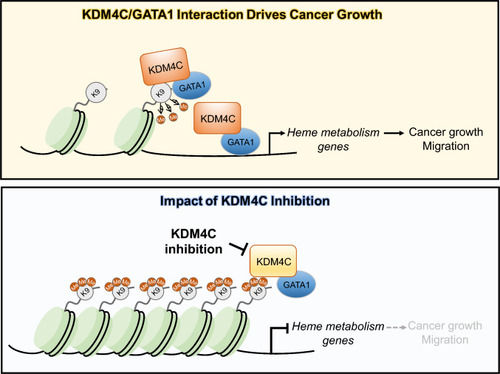
Schematic illustration of KDM4C-GATA1 axis in heme metabolism and cancer progression. In the active state (top panel), KDM4C interacts with GATA1, demethylating H3K9me3 marks at heme metabolism gene promoters such as |