- Title
-
How Tumors Affect Hemodynamics: A Diffusion Study on the Zebrafish Transplantable Model of Medullary Thyroid Carcinoma by Selective Plane Illumination Microscopy
- Authors
- Carra, S., Gaudenzi, G., Franceschetti, G., Collini, M., Sironi, L., Bouzin, M., Persani, L., Chirico, G., Vitale, G., D'Alfonso, L.
- Source
- Full text @ Int. J. Mol. Sci.
|
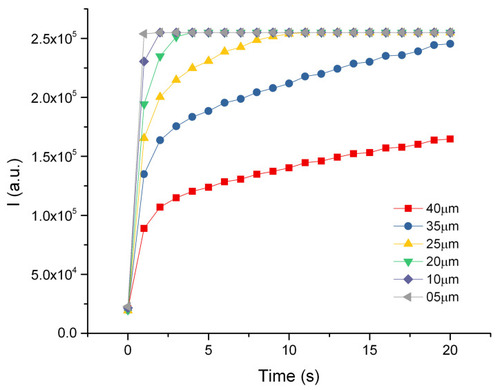
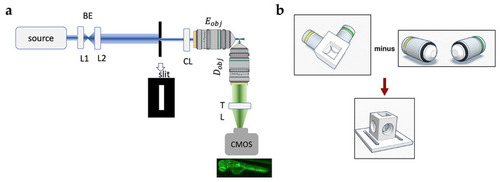
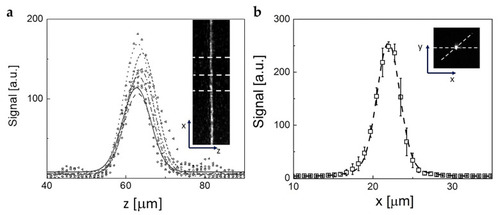
Analysis of the time stack of images acquired for 5 min (5 s time steps) after the injection of very concentrated rhodamine in solidified agarose. ( |
|
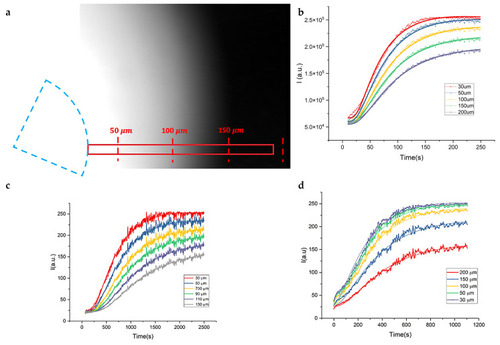
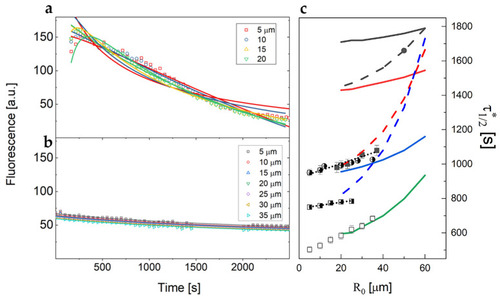
Intensity profiles of diffusing rhodamine in jellified 1% agarose solution. Different intensity profiles are recorded at different distances from the injection point as a function of time. |
|
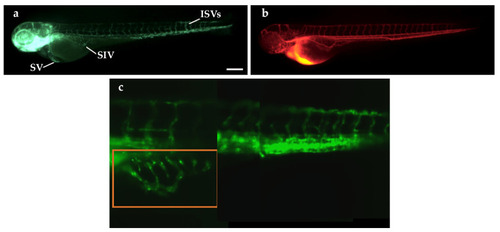
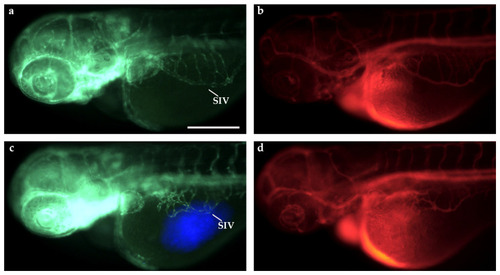
Microangiography assay in |
|
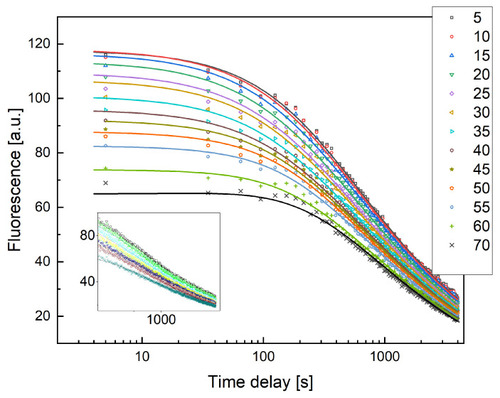
Fluorescent dextran intensity profiles versus time measured at increasing distances from the SIV plexus for a control zebrafish embryo, as shown in the legend. Solid lines are the best fit of the data to Equation (4), performed to determine the value of the diffusion coefficient D, kept as a shared global parameter. The time delay between the injection and the observation was set to |
|
Microangiography assays in TT-xenografted and control |
|
Examples of fluorescent dextran intensity profiles over time measured on different xenografted zebrafish samples at different distances from the injection point. Solid lines are the best fit with |
|
( |
|
( |
|
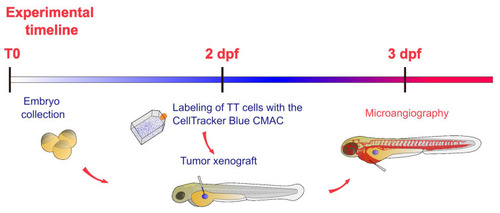
Schematic representation of the experimental timeline of experiments in zebrafish embryos. After the collection (T0), zebrafish embryos were incubated at 28 °C up to 2 dpf. At this stage labeled TT cells were implanted in embryos. After the xenograft, embryos were incubated at 32 °C for 24 h. The day after, microangiography assays were performed on 3 dpf embryos. |