- Title
-
Inhibition of PNCK inflames tumor microenvironment and sensitizes head and neck squamous cell carcinoma to immune checkpoint inhibitors
- Authors
- Ding, Q., Weng, Y., Li, Y., Lin, W., Lin, X., Lin, T., Yang, H., Xu, W., Wang, J., Ying, H., Qiu, S.
- Source
- Full text @ J Immunother Cancer
|
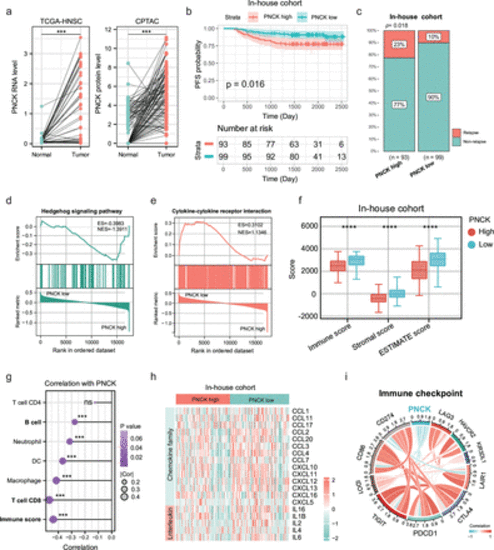
An initial exploration into the predictive significance of PNCK for prognosis and immune-inflammatory response. (a) Differential expression of PNCK in TCGA-HNSC cancer tissues and normal tissues. Two-tailed paired Student’s t-test. (b) Kaplan-Meier progression-free survival (PFS) curve for 193 head and neck tumor patients with high or low PNCK expression in the cohort from Fujian Provincial Cancer Hospital. Two-sided log-rank test. (c) Recurrence rates in patients from Fujian Provincial Cancer Hospital with high versus low PNCK expression. The red bar represents relapse, and the blue bar indicates non-relapse. Two-tailed χ2 test. (d, e) GSEA revealing pro-carcinogenic-related and immune-related pathways correlated with PNCK expression in the in-house dataset (n=193 biologically independent samples). High expression of PNCK was enriched in the Hedgehog signaling pathway (d), whereas low expression was enriched in the cytokine-cytokine receptor pathway (e). (f) Significant differences in immune score, stromal score, and ESTIMATE score between PNCK high and low expression groups. The upper, middle and lower horizontal lines of the box represent the upper, median and lower quartile respectively. Two-tailed unpaired Student’s t-test. Data are presented as mean values±SD. (g) Association of PNCK expression with diverse immune cells and the immune score. Two-tailed Spearman correlation is reported. (h) The heatmap illustrates the differential cytokine expression profiles between high and low PNCK expression groups. (i) Association between PNCK expression and immune checkpoint marker expression. Two-tailed Spearman correlation is reported. HNSC, head and neck squamous cell. *** indicates p < 0.001, and **** indicates p < 0.0001. |
|
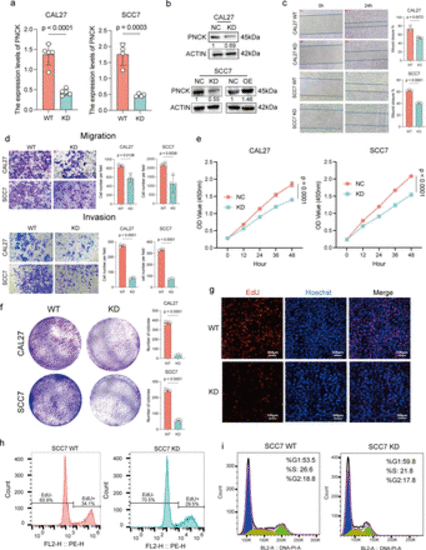
Cellular-level validation of PNCK promoting malignant biological behaviors in HNSCC. (a) RT-qPCR verification of PNCK knockdown efficiency. A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. (b) Western blot verification of PNCK knockdown and overexpression efficiency. The numbers below each panel represent the relative protein expression, with the NC group set to 1. (c) A scratch assay was performed to assess cell migration. The wound healing was monitored and quantified using a two-tailed unpaired Student’s t-test. Data are presented as mean values±SD. (d) Transwell migration and invasion assays were conducted to evaluate the migratory and invasive capabilities of HNSCC cells with altered PNCK expression, using a two-tailed unpaired Student’s t-test. Data are presented as mean values±SD. (e) Cell proliferation was assessed using the CCK8 assay. Two-way ANOVA with Tukey’s multiple comparison test. Data are presented as mean values±SD. (f) The ability of HNSCC cells to form colonies was evaluated using a colony formation assay, using a two-tailed unpaired Student’s t-test. Data are presented as mean values±SD. (g) EdU incorporation assay was performed to detect proliferating cells. Blue represents cell nuclei stained with Hoechst, and red represents EdU-positive cells; scale bar: 100 µm. (h) Flow cytometry was used to quantify EdU-positive cells. (i) The cell cycle distribution of HNSCC cells with altered PNCK expression was analyzed by flow cytometry. ANOVA, analysis of variance; HNSCC, head and neck squamous cell carcinoma. |
|
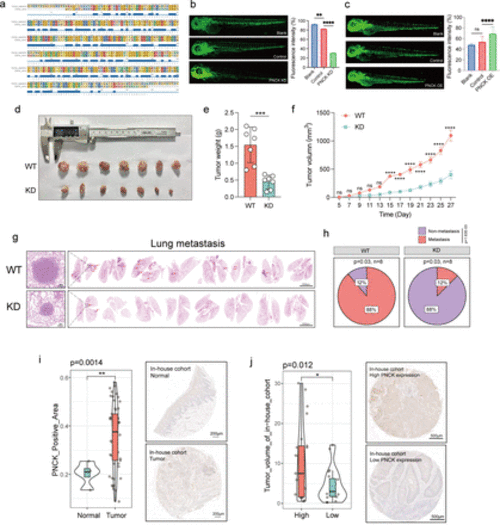
Knockdown of PNCK significantly inhibits in vivo growth of HNSCC. (a) Homology comparison of PNCK gene between zebrafish and humans. The squares and letters in various colors at the top represent different amino acid sequences, while the blue squares at the bottom indicate overlapping portions of these sequences. (b, c) Comparison of zebrafish blood vessel fluorescence intensity after PNCK knockdown (b) and overexpression (c).A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. (d) Subcutaneous tumor growth in C3H mice (n=7 biologically independent samples). (e) Comparison of mouse tumor weights. A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. (f) Mouse tumor growth curve. A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. (g) HE staining of lung metastatic nodules in mice (n=8 biologically independent samples). (h) The pie chart shows the percentage of mice that developed pulmonary metastases. Two-tailed χ2 test. (i) Difference in PNCK-positive area and HE staining between normal and tumor tissues in HNSCC tissue microarray (n=60 biologically independent samples). A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. (j) Difference in tumor volume between PNCK high and low expression groups in HNSCC tissue microarray, and corresponding HE staining (n=60 biologically independent samples). A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. HNSCC, head and neck squamous cell carcinoma. * indicates p < 0.05, ** indicates p < 0.01, *** indicates p < 0.001, and **** indicates p < 0.0001. |
|
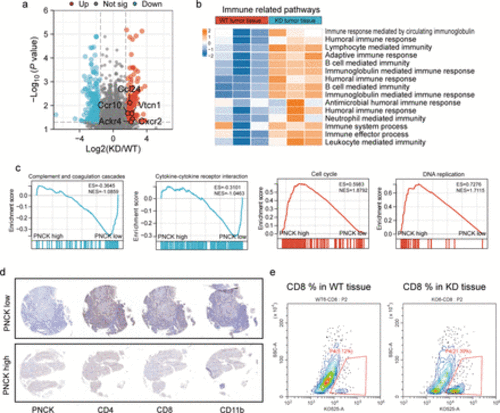
Knockdown of the Pnck gene induces antitumor immune response. (a) Volcano plot showing DEGs in the sequencing results, including upregulation of genes encoding immune cell chemotactic factors, like Ccl24, Ccr10, Vtcn1, Ackr4 and Cxcr2. Differentially expressed genes were identified with the threshold of |log2(fold change)| >1 and false discovery rate (FDR)<0.05. (b) Heatmap showing the enrichment and upregulation of immune-related pathways in the Pnck-KD group. (c) GSEA enrichment analysis of complement and cytokine-related pathways in the PNCK low-expression group, and GSEA enrichment of cell cycle and DNA repair-related pathways in the high-expression group. (d) Immunohistochemistry demonstrated the expression levels of CD4, CD8, and CD163 in PNCK high and low groups. (e) Infiltration abundance of CD8 T cells in mouse tumors in the WT and Pnck-KD group (n=3 biologically independent samples). |
|
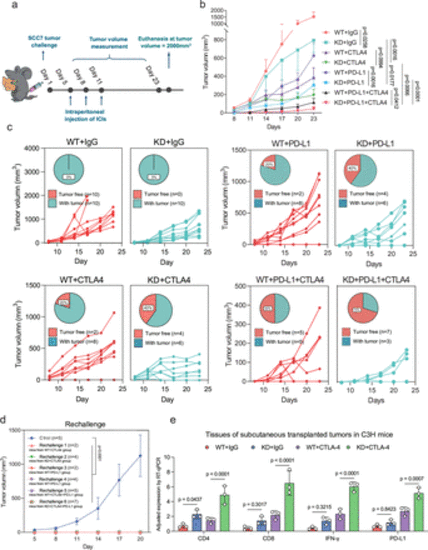
PNCK knockdown enhances the efficacy of immune checkpoint inhibition therapy. (a) The experimental schedule for the SCC7 cell challenge in the C3H mice tumor model is as follows. Intraperitoneal injections of ICIs were administered at a dose of 200 µg per mouse on the 5th, 8th, and 11th days after tumor implantation. The experiment was terminated on the 23rd day, and the tumors were removed for subsequent analysis. If the tumor volume reached 2000 mm³ during the experiment, euthanasia was performed immediately. (b) Overall tumor growth curves in different groups of mice. Mice were intraperitoneally treated with 200 µg anti-PD-L1 or 200 µg anti-CTLA4 on days 5, 8 and 11 after tumor inoculation. A rat IgG isotype antibody was applied as a control. Two-way ANOVA with Tukey’s multiple comparison test. Data are presented as mean values±SEM. The statistical comparisons among these groups were listed as follows: WT+IgG vs KD+IgG, p=0.0258; WT+CTLA4 vs KD+CTLA4, p=0.0994; WT+PD-L1 vs KD+PD-L1, p=0.0016; WT+CTLA4 + PD-L1 vs KD+CTLA4 + PD-L1, p=0.0412; KD+IgG vs KD+CTLA4, p=0.0016; KD+IgG vs KD+PD-L1, p=0.0177; KD+CTLA4 vs KD+CTLA4 + PD-L1, p=0.0066; KD+PD-L1 vs KD+CTLA4 + PD-L1, p<0.0001. (c) Individual tumor growth curves and percentages in different groups of mice. (d) Tumor growth curve in mice with cured tumors after subcutaneous reimplantation of HNSCC. Two-way ANOVA with Tukey’s multiple comparison test was performed. Data are presented as mean values±SD. (e) Expression levels of immune markers and cytokines in tumors of mice in different groups. Kruskal-Wallis with Dunn’s multiple comparison test are reported. ANOVA, analysis of variance; ICIs, immune checkpoint inhibitors. |
|
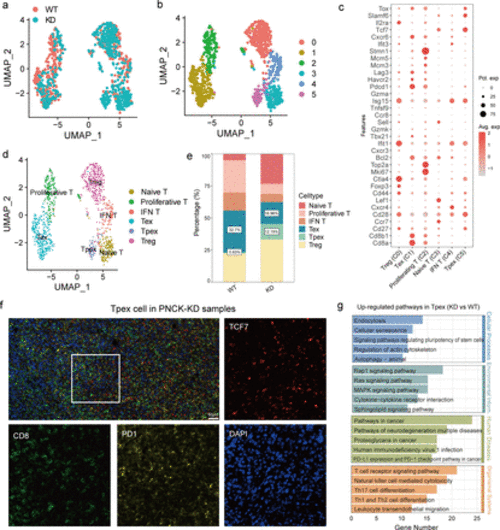
Heterogeneity analysis of T cell subtypes. (a, b) UMAP analysis shows the distribution of T cell subtypes in different samples. Each point represents a single cell, colored by its subtype. (c) Bubble plots showing the expression of commonly used marker genes for each T cell subtype, used for annotating cell subtypes. The size of the bubble represents the percentage of cells expressing the marker gene, and the color intensity indicates the expression level. (d) UMAP analysis shows the distribution of different T cell subtypes. Different colors represent different T cell subtypes. (e) Proportion of each T cell subtype in the WT and knockdown groups. Bar graphs illustrate the percentage of each subtype in the two groups. (f) Multiplex immunofluorescence probed the content of Tpex cells (TCF7+ PD1+ CD8+) in PNCK-KD samples. (g) KEGG functional enrichment analysis of DEGs in the Tpex cell subtype. The bar graph shows the top enriched pathways, with the length of the bar indicating the enrichment score. DEGs, differentially expressed genes. |
|
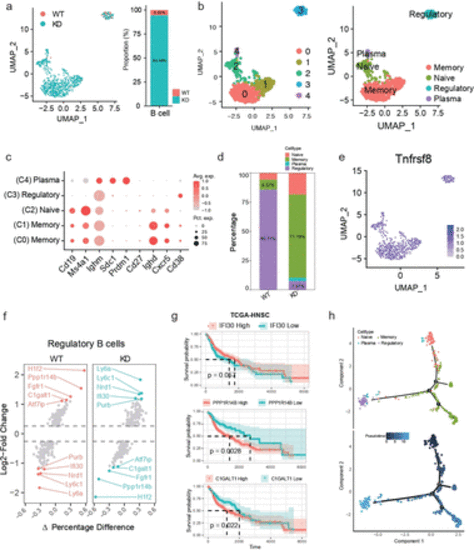
Heterogeneity analysis of B cell subtypes. (a) UMAP analysis shows the distribution of B cells in different groups. Each point represents a single cell, colored by its subtype. (b) UMAP analysis shows the distribution of B cell subtypes in different samples. Each point represents a single cell. Different colors represent different B cell subtypes. (c) Bubble plots showing the expression of commonly used marker genes for each B cell subtype, used for annotating cell subtypes. The size of the bubble represents the percentage of cells expressing the marker gene, and the color intensity indicates the expression level.(d) Proportion of each B cell subtype in the WT and knockdown groups. (e) Expression of the B cell marker Tnfrsf8 (Cd30). (f) Differential genes in regulatory B cells between different groups. (g) Prognostic value of specific genes in regulatory B cells, based on survival analysis. A Two-tailed log-rank test was performed (h) Pseudotime analysis shows the developmental trajectory of B cell subtypes. |
|
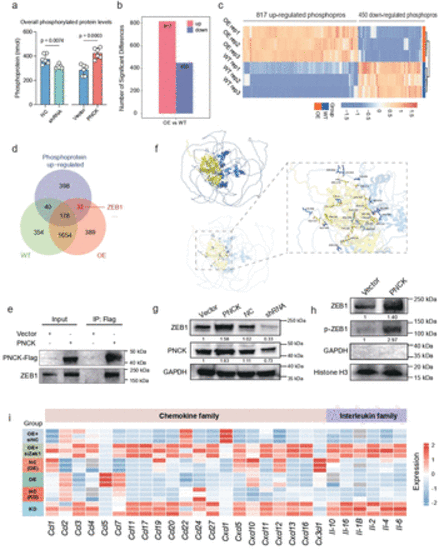
PNCK regulates ZEB1 nuclear translocation and reshapes the tumor immune-suppressive microenvironment. (a) Quantification of overall phosphorylated protein levels in various cell samples to assess changes in the phosphorylation status. A two-tailed unpaired Student’s t-test was performed. Data are presented as mean values±SD. (b) Differential phosphorylated proteins between the PNCK overexpression group and the WT group. (c) Heatmap showing the expression pattern of differentially phosphorylated proteins in the PNCK OE and WT groups. (d) Venn diagram showing the intersection of upregulated phosphorylated proteins and proteins interacting with PNCK in WT and OE groups. (e) Co-immunoprecipitation (CO-IP) validation demonstrates the interaction between PNCK and ZEB1. (f) Surface diagram of the docking model and their interfacing residues between ZEB1 and PNCK protein (ZEB1, blue; PNCK, yellow; hydrogen bond interaction, dotted line). (g) Western blot analysis shows changes in ZEB1 expression levels in response to alterations in PNCK expression. (h) Western blot analysis shows the upregulation of ZEB1 and phosphorylated ZEB1 (p-ZEB1) in the nuclear protein fraction of the PNCK OE group. GAPDH was used as a cytoplasmic protein loading control, and Histone H3 was used as a nuclear protein loading control. (i) Cytokine array analysis of numerous immune-related cytokines across multiple experimental groups, including KD, NC(KD), OE, NC(OE), OE+siZEB1, and OE+siNC. |