- Title
-
Effect of nonsense-mediated mRNA decay factor SMG9 deficiency on premature aging in zebrafish
- Authors
- Lai, S., Shiraishi, H., Sebastian, W.A., Shimizu, N., Umeda, R., Ikeuchi, M., Kiyota, K., Takeno, T., Miyazaki, S., Yano, S., Shimada, T., Yoshimura, A., Hanada, R., Hanada, T.
- Source
- Full text @ Commun Biol
|
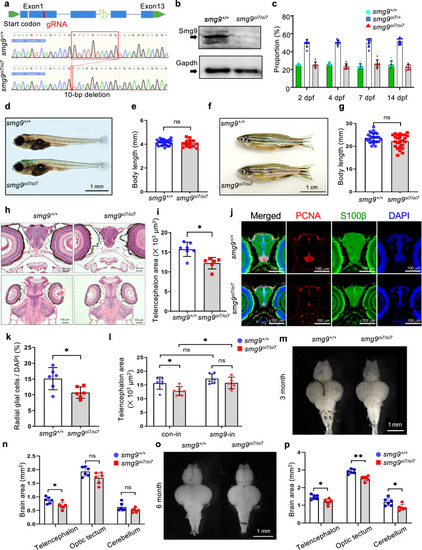
Smg9-deficient zebrafish display brain malformations. |
|
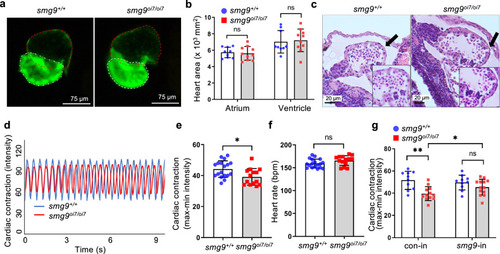
Impaired cardiac contractions in Smg9-deficient zebrafish. |
|
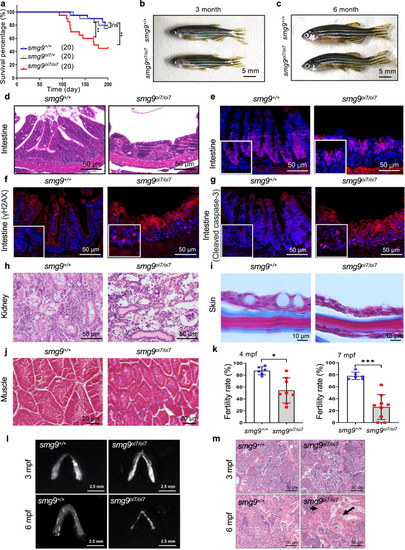
Premature aging phenotype in various tissues in Smg9-deficient zebrafish. |
|
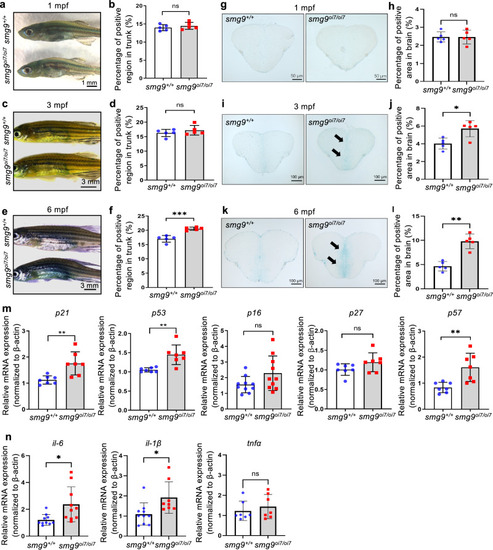
Age-related staining and markers in Smg9-deficient zebrafish. |
|
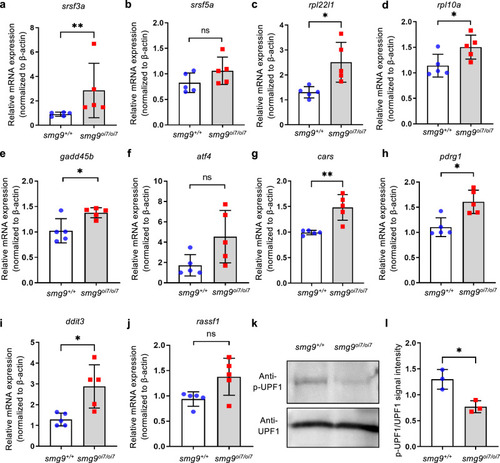
Upregulation of endogenous targets of nonsense-mediated mRNA decay (NMD) in Smg9-deficient zebrafish. |
|
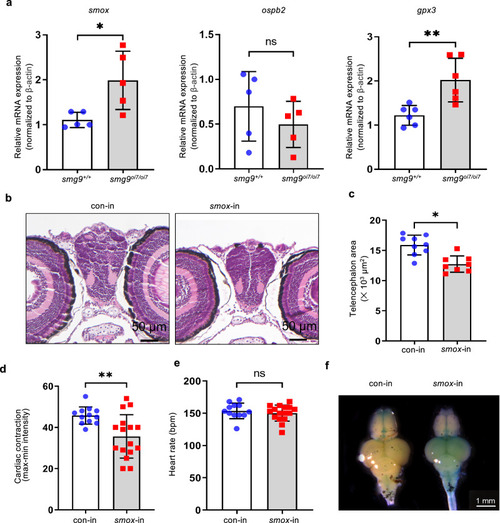
Smox overexpression is sufficient for brain and heart defects in Smg9-deficient zebrafish. |
|
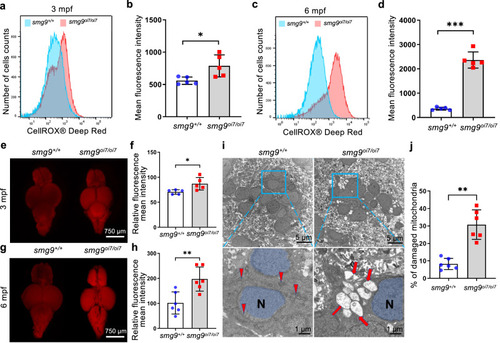
ROS and acrolein accumulation in Smg9-deficient zebrafish. |
|
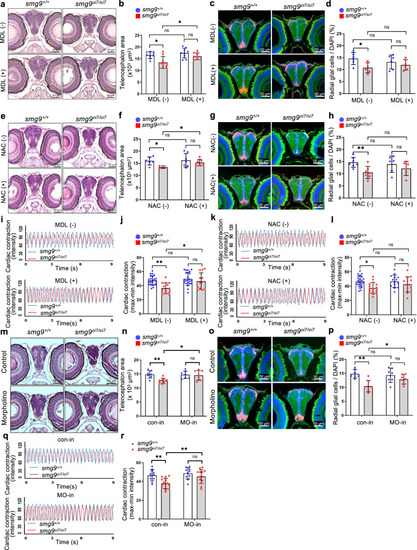
Smox is required for brain and heart defects in Smg9-deficient zebrafish. |