- Title
-
Rare coding variants in NOX4 link high ROS levels to psoriatic arthritis mutilans
- Authors
- Wang, S., Nikamo, P., Laasonen, L., Gudbjornsson, B., Ejstrup, L., Iversen, L., Lindqvist, U., Alm, J.J., Eisfeldt, J., Zheng, X., Catrina, S.B., Taylan, F., Vaz, R., Ståhle, M., Tapia-Paez, I.
- Source
- Full text @ EMBO Mol. Med.
|
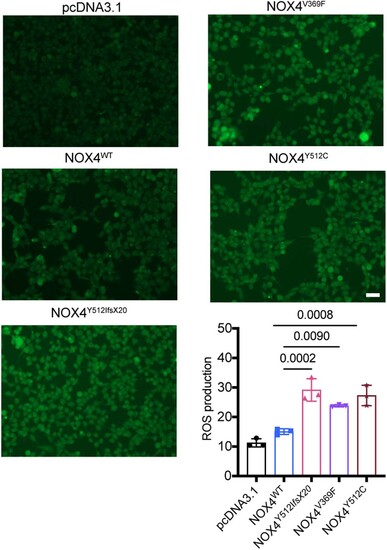
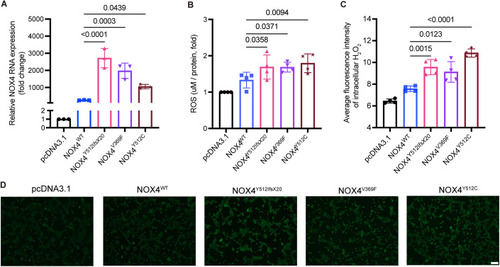
Elevated ROS levels in HEK293 stable transfected cell lines expressing NOX4 variants. HEK293 cells were subjected to 12 h of serum starvation after stably transfection with following plasmids: pcDNA3.1, NOX4wt, NOX4Y512IfsX20, NOX4Y512C, and NOX4V369F. Fluorescence imaging was conducted following the incubation with 10 μM DCFH-DA. Representative photomicrographs of the fluorescence are displayed and quantification of the mean fluorescence intensity was performed with ImageJ software. N = 3. Scale bars: 100 μm. Data information: DCFH-DA 2′,7′-dichlorofluorescein diacetate. Data presented as mean ± SD. N = biological replicates. The P value was calculated by the ordinary one-way ANOVA multiple comparisons with Turkey correction of multiple hypothesis tests. Source data are available online for this figure. |
|
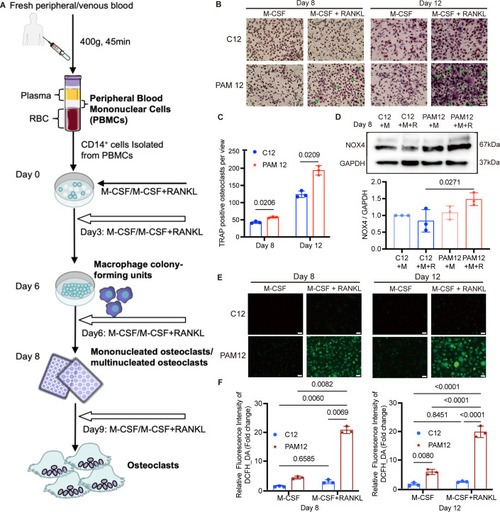
Impact of ROS on osteoclasts differentiation from PsA961 (NOX4Y512C). (A) Cells were cultured in the presence of M-CSF or M-CSF and RANKL. Visualization of ROS detected by DCFH-DA on Day 6 and Day 11, indicating a higher ROS effect in PsA961-differentiated osteoclasts. Scale bar: 100 μm. (B) The relative fluorescence intensity of ROS was quantified by GraphPad Prism9.0.0. N = 3. Data information: DCFH-DA 2′,7′-dichlorofluorescein diacetate. N = biological replicates. For graph (B), error bars in figure represent mean ± SD (two-way ANOVA with Tukey’s multiple comparisons tests). Source data are available online for this figure. |
|
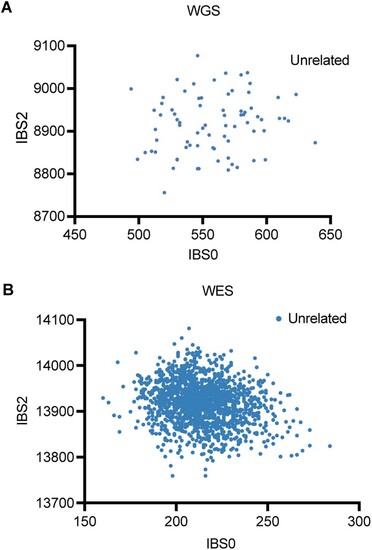
Relatedness plot for WGS (A) and WES (B) samples. (A) Relatedness plot for WGS samples. Six samples from the SweGene database (https://swefreq.nbis.se/) were added to the WGS analysis. Each dot represents a pair of samples. (B) Relatedness analysis of all WES samples. Data information: IBS0 is the number of sites where 1 sample is homozygous for the reference allele and the other is homozygous for the alternate allele. IBS2, is the count of sites where a pair of samples were both homozygous or both heterozygous. WGS Whole-Genome Sequencing, WES Whole-Exome Sequencing. Source data are available online for this figure. |
|
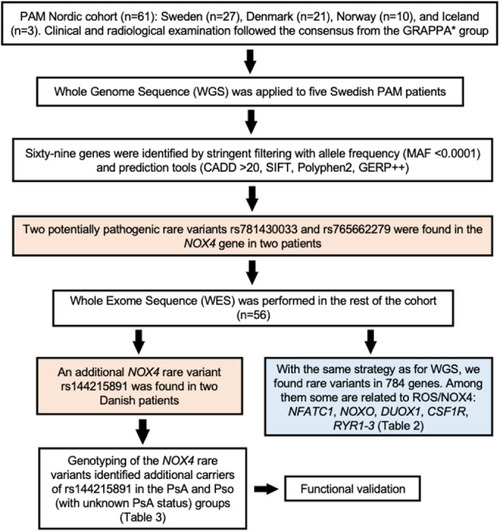
Flowchart outlining the study design and the criteria applied in the filtering of rare variants found in PAM patients by next-generation sequencing. Data information: *GRAPPA Group for Research and Assessment of Psoriasis and Psoriatic Arthritis. |
|
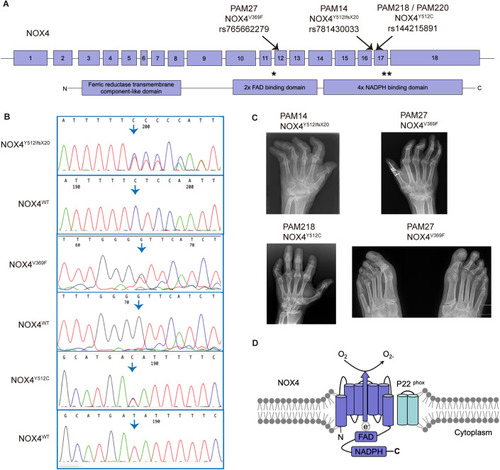
Next-generation sequencing reveals rare variants in the ( |
|
HEK293 cell lines overexpressing the rare variants found in ( |
|
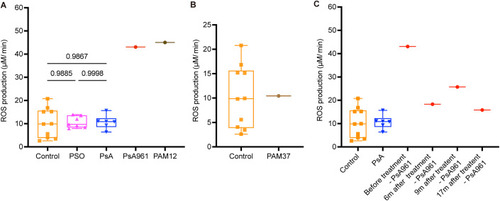
Electron paramagnetic resonance (EPR) shows increased levels of ROS in the patients PAM12 and PsA961 and the impact of anti-TNF-α treatment in PsA961. ( |
|
PAM12-derived osteoclasts show enhanced differentiation and increased ROS generation activity compared to osteoclast-derived cells from healthy control. ( |
|
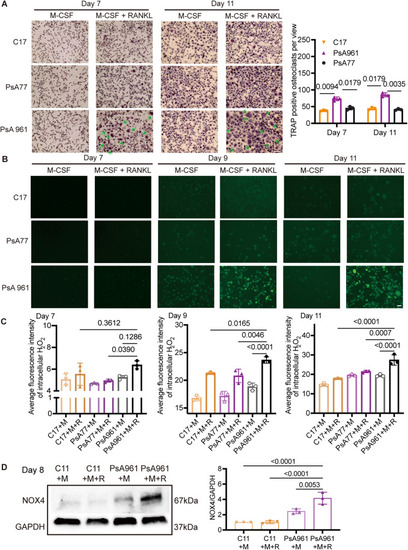
PsA961 ( ( |
|
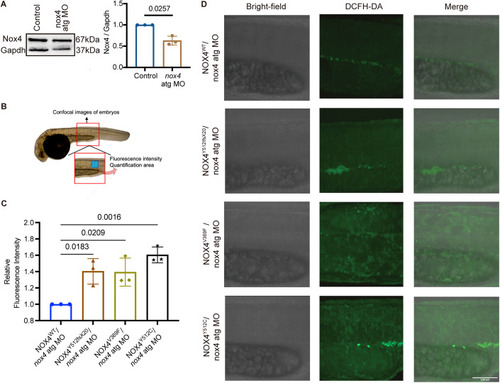
ROS production is increased in zebrafish embryos injected with ( |