- Title
-
Analysis of SMAD1/5 target genes in a sea anemone reveals ZSWIM4-6 as a novel BMP signaling modulator
- Authors
- Knabl, P., Schauer, A., Pomreinke, A.P., Zimmermann, B., Rogers, K.W., Čapek, D., Müller, P., Genikhovich, G.
- Source
- Full text @ Elife
|
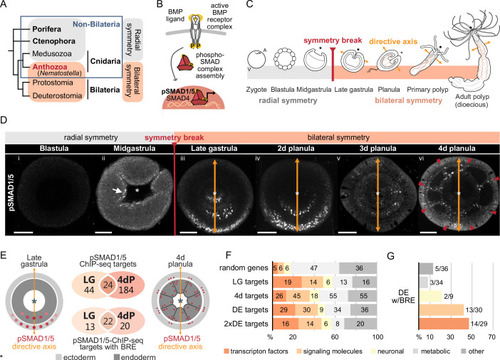
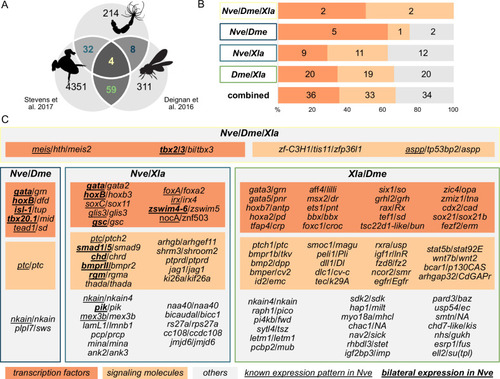
Bilateral body symmetry of the non-bilaterian sea anemone Nematostella is BMP signaling-dependent. (A) Bilateral body symmetry is observed in Bilateria and in anthozoan Cnidaria. (B) BMP signaling is initiated by BMP ligands binding to BMP receptors that trigger phosphorylation, assembly, and nuclear translocation of a pSMAD1/5/SMAD4 complex to regulate gene expression. (C) A BMP signaling-dependent symmetry break at late gastrula (LG) stage results in the formation of the secondary (directive) body axis in the sea anemone Nematostella. (D) BMP signaling dynamics during Nematostella development. No pSMAD1/5 is detectable in the blastula (Di). Nuclear pSMAD1/5 is localized in the blastopore lip of midgastrula (Dii), forms a gradient along the directive axis in the LG (Diii) and 2d planula (Div). By day 3, the gradient progressively disperses (Dv), and the signaling activity shifts to the eight forming endodermal mesenteries (Dvi) and to the ectodermal stripes vis-à-vis the mesenteries (arrowheads). Images (Dii–Dvi) show oral views (asterisks). Scale bars 50 µm. (E) Comparison of the direct BMP signaling targets at LG and 4dP shows little overlap. Schemes show oral views of a LG and a 4dP with red spots indicating the position of pSMAD1/5-positive nuclei in the ectoderm (light gray) and endoderm (dark gray). (F) Transcription factors, signaling molecules, and neuronal genes are overrepresented among the pSMAD1/5 targets compared to the functional distribution of 100 random genes. LG, late gastrula targets; 4dP, 4d planula targets; DE, pSMAD1/5 ChIP targets differentially expressed in BMP2/4 and/or GDF5-like morphants (padj≤0.05); 2xDE targets, pSMAD1/5 ChIP targets differentially expressed in BMP2/4 and/or GDF5-like morphants (padj≤0.05) showing ≥2-fold change in expression. (G) Fractions of each functional category of the differentially expressed pSMAD1/5 target genes (see panel F) containing BMP response elements (BREs). |
|
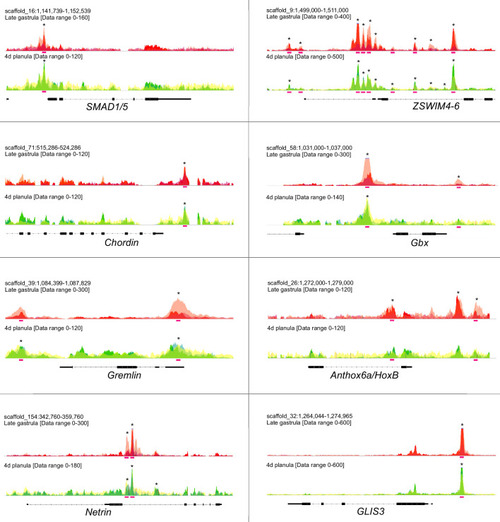
Sequencing coverage profile shows the enrichment of pSMAD1/5 binding at the target genes. Overlays of the three biological replicates are shown. Statistically significant peaks are marked with asterisks. Pink bars, BMP response elements (BREs). |
|
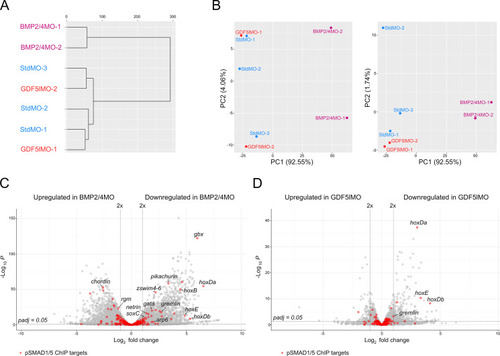
Transcriptomic comparison of BMP2/4, GDF5-like, and control morphants and differential expression of pSMAD1/5 ChIP targets upon different knockdowns. ( |
|
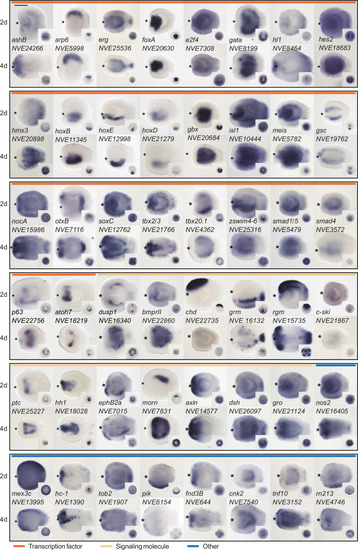
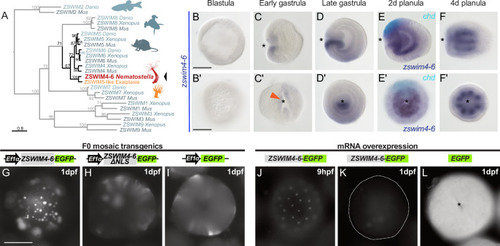
Expression patterns of a selection of the direct targets of BMP signaling in 2d and 4d planulae. In lateral views, the oral end is marked with an asterisk, inlets show oral views. Scale bar 100 µm. |
|
( |
|
( |
|
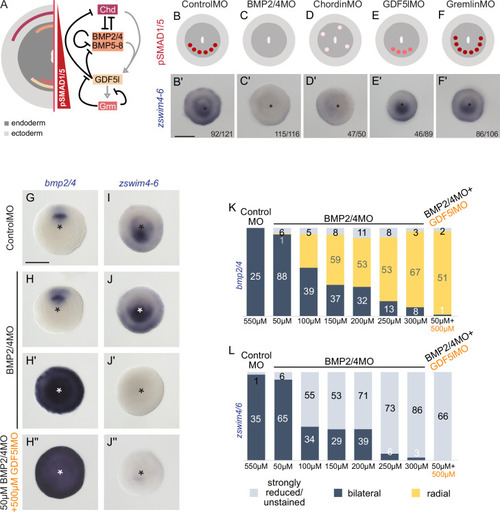
zswim4-6 reacts differently to the downregulation of BMP2/4 and GDF5-like. (A) Expression domains and putative interactions of the BMP network components in 2d embryos (based on Genikhovich et al., 2015). (B–F’) Effect of the knockdown (KD) of the individual BMP signaling components on nuclear pSMAD1/5 and on zswim4-6 expression. zswim4-6 expression is abolished upon KD of bmp2/4 and chordin, but not in KD of gdf5-l and gremlin. Sketches in (B–F) show that the gradient is lost upon BMP2/4 and Chd KD, reduced upon GDF5-like KD and expanded upon Gremlin KD (based on Genikhovich et al., 2015). (G–L) KD of bmp2/4 using different concentrations of BMP2/4MO results in either normal, bilaterally symmetric marker expression or complete radialization but no intermediate phenotypes. Instead, the penetrance of the radialization phenotype increases with the increase of the BMP2/4MO concentration. Co-injection of the lowest, ineffective BMP2/4MO concentration with GDF5lMO also results in a complete radialization. All stainings are performed on 2d planulae. Scale bars 100 µm. |
|
Two type I BMP receptors display partial redundancy in regulating Expression of BMP receptors ( |
|
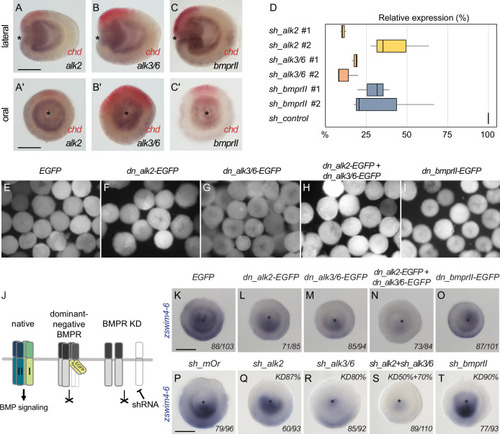
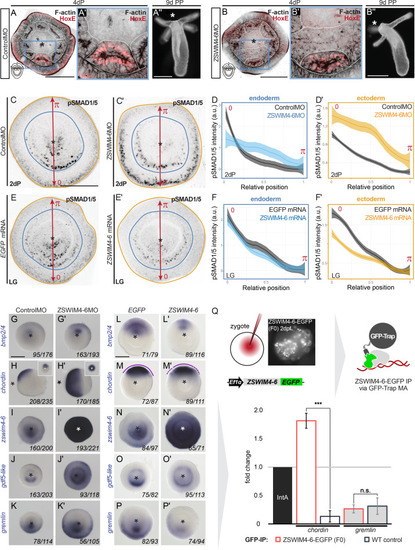
ZSWIM4-6 is a modulator of BMP signaling that appears to act as transcriptional repressor. (A–B’’) Morpholino knockdown (KD) of zswim4-6 results in patterning defects. (A) and (B) show confocal sections across the pharyngeal region of 4d planulae stained with antiHoxE antibody (Genikhovich et al., 2015) and phalloidin. (A’) and (B’) show the areas boxed in (A) and (B). White dotted lines delineate mesenterial chambers. In the 4d planula, the HoxE-positive mesenterial chamber does not reach the pharynx, which leads to the fusion of neighboring chambers (compare A, A’ with B, B’). This results in the formation of three instead of four tentacles in the 9d polyp (compare A’’ with B’’). (C–F’) Immunofluorescence and quantification of relative nuclear anti-pSMAD1/5 staining intensities in 2d ZSWIM4-6 morphants (C–D’) and upon zswim4-6 mRNA overexpression in the late gastrula (E–F’). Intensity measurements (arbitrary units, a.u.) are plotted as a function of the relative position of each nucleus in the endoderm or in the ectoderm along a 180° arc from 0 (high signaling side) to π (low signaling side). The measurements from Control MO embryos (n = 10) and ZSWIM4-6MO embryos (n = 10), as well as egfp mRNA embryos (n = 22 for the endodermal, and n = 24 for the ectodermal measurements) and zswim4-6 mRNA embryos (n = 8 for the endodermal, and n = 9 for the ectodermal measurements) are described by a LOESS smoothed curve (solid line) with a 99% confidence interval for the mean (shade). For visualization purposes, the intensity values were normalized to the upper quantile value among all replicates and conditions of each control-experiment pair. (G–K’) Expression of zswim4-6 and BMP network components in the 2d planula upon morpholino KD of zswim4-6; All images except for (H) and (H’) show oral views. (L–P’) Expression of zswim4-6 and BMP network components in late gastrula (30 hr) upon zswim4-6 mRNA injection; oral views, purple dashed line marks the loss of a sharp boundary of chd expression. In (A–P’), asterisks mark the oral side. In (A-P’), scale bars 100 µm. (Q) ChIP with GFP-Trap detects ZSWIM4-6-EGFP fusion protein in the vicinity of the pSMAD1/5 binding site in the upstream regulatory region of chordin but not of gremlin. Experiment on biological quadruplicates. Mean enrichments and standard deviations are shown. |
|
Testing ZSWIM4/6 morpholino specificity. ( |
|
( |
|
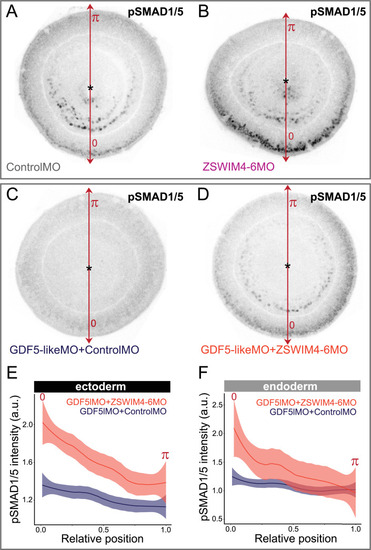
ZSWIM4-6 knockdown partially rescues the reduction of pSMAD1/5 caused by GDF5-like knockdown. Oral views of 2 dpf embryos stained for pSMAD1/5. ( |
|
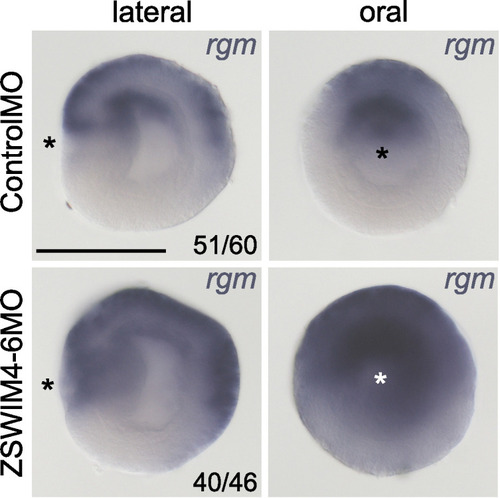
In situ hybridization on 2d planulae. Asterisks denote the location of the blastopore. Scale bar 100 µm. |
|
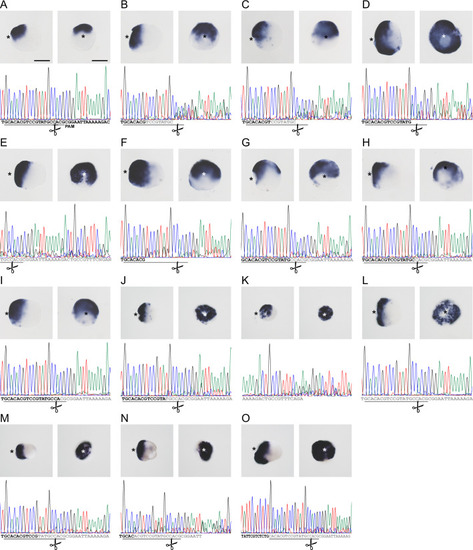
CRISPR/Cas9-mediated mutagenesis of ( |