Figure 1—figure supplement 1.
- ID
- ZDB-FIG-240211-32
- Publication
- Knabl et al., 2024 - Analysis of SMAD1/5 target genes in a sea anemone reveals ZSWIM4-6 as a novel BMP signaling modulator
- Other Figures
-
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 2.
- Figure 2.
- Figure 3.
- Figure 4.
- Figure 5—figure supplement 1—source data 1.
- Figure 5—figure supplement 1—source data 1.
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 2—source data 1.
- Figure 6—figure supplement 3—source data 1.
- Figure 6—figure supplement 4.
- Figure 6—figure supplement 5.
- All Figure Page
- Back to All Figure Page
|
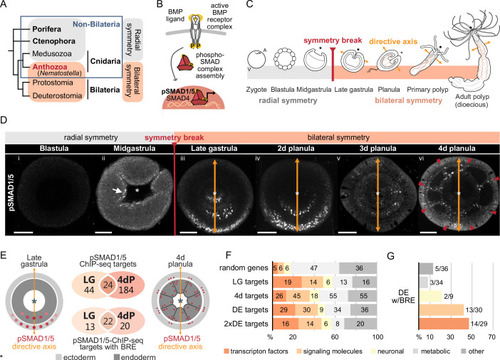
Bilateral body symmetry of the non-bilaterian sea anemone Nematostella is BMP signaling-dependent. (A) Bilateral body symmetry is observed in Bilateria and in anthozoan Cnidaria. (B) BMP signaling is initiated by BMP ligands binding to BMP receptors that trigger phosphorylation, assembly, and nuclear translocation of a pSMAD1/5/SMAD4 complex to regulate gene expression. (C) A BMP signaling-dependent symmetry break at late gastrula (LG) stage results in the formation of the secondary (directive) body axis in the sea anemone Nematostella. (D) BMP signaling dynamics during Nematostella development. No pSMAD1/5 is detectable in the blastula (Di). Nuclear pSMAD1/5 is localized in the blastopore lip of midgastrula (Dii), forms a gradient along the directive axis in the LG (Diii) and 2d planula (Div). By day 3, the gradient progressively disperses (Dv), and the signaling activity shifts to the eight forming endodermal mesenteries (Dvi) and to the ectodermal stripes vis-à-vis the mesenteries (arrowheads). Images (Dii–Dvi) show oral views (asterisks). Scale bars 50 µm. (E) Comparison of the direct BMP signaling targets at LG and 4dP shows little overlap. Schemes show oral views of a LG and a 4dP with red spots indicating the position of pSMAD1/5-positive nuclei in the ectoderm (light gray) and endoderm (dark gray). (F) Transcription factors, signaling molecules, and neuronal genes are overrepresented among the pSMAD1/5 targets compared to the functional distribution of 100 random genes. LG, late gastrula targets; 4dP, 4d planula targets; DE, pSMAD1/5 ChIP targets differentially expressed in BMP2/4 and/or GDF5-like morphants (padj≤0.05); 2xDE targets, pSMAD1/5 ChIP targets differentially expressed in BMP2/4 and/or GDF5-like morphants (padj≤0.05) showing ≥2-fold change in expression. (G) Fractions of each functional category of the differentially expressed pSMAD1/5 target genes (see panel F) containing BMP response elements (BREs). |