- Title
-
PFHxS Exposure and the Risk of Non-Alcoholic Fatty Liver Disease
- Authors
- Ulhaq, Z.S., Tse, W.K.F.
- Source
- Full text @ Genes (Basel)
|
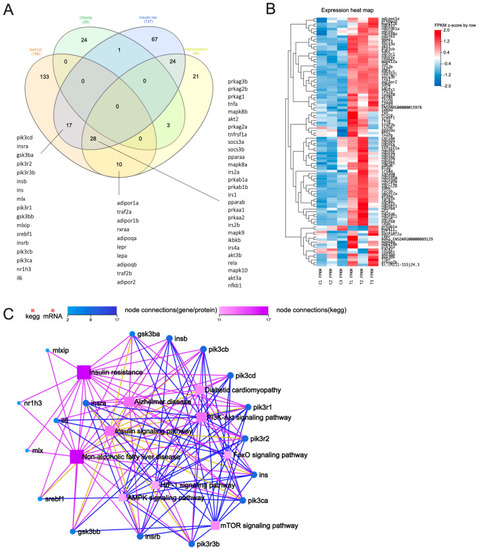
Transcriptomics analysis of 5-dpf zebrafish larvae exposed to 5 μM PFHxS. ( PHENOTYPE:
|
|
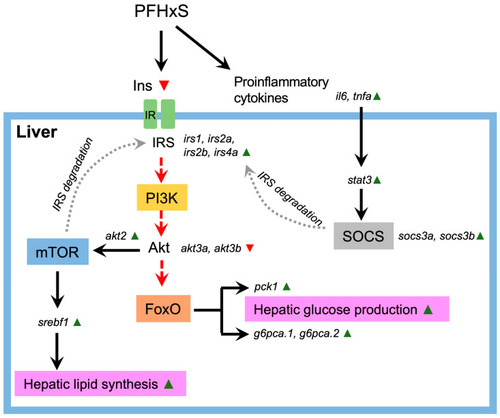
Exposure to PFHxS triggers a cascade of events leading to NAFLD. Pro-inflammatory cytokines, activated in response to PFHxS exposure, initiate the ubiquitin-mediated proteasomal degradation of insulin receptor substrates 1 and 2 (IRS1/2), essential molecules for insulin transduction, via the phosphoinositide 3-kinase (PI3K)/Akt pathway. This degradation is facilitated through the activation of the suppressor of cytokine signaling (SOCS) by the signal transducer and activator of transcription 3 (STAT3). Alternatively, the proteasomal degradation of IRS1/2 can promote mTOR signaling. The depletion of IRS1/2 disrupts Akt signaling, impairing Akt-mediated phosphorylation and leading to the further ubiquitin-mediated proteasomal degradation of forkhead box protein O (FoxO). FoxO activation subsequently upregulates gluconeogenic enzymes such as phosphoenolpyruvate carboxykinase (PCK1) and glucose-6-phosphatase (G6P), resulting in increased hepatic glucose production. Conversely, the PFHxS activation of |
|
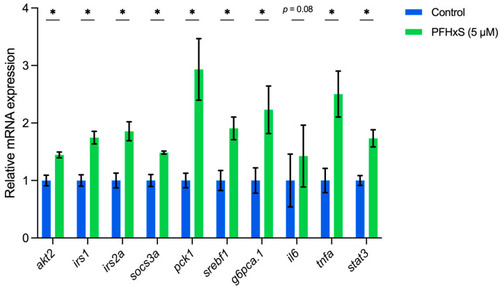
The mRNA expression levels of 10 selected genes ( |