- Title
-
Phosphorylation of Def Regulates Nucleolar p53 Turnover and Cell Cycle Progression through Def Recruitment of Calpain3
- Authors
- Guan, Y., Huang, D., Chen, F., Gao, C., Tao, T., Shi, H., Zhao, S., Liao, Z., Lo, L.J., Wang, Y., Chen, J., Peng, J.
- Source
- Full text @ PLoS Biol.
|
Wild-type p53, but not the mutant p53R175H, is a substrate of CAPN3. |
|
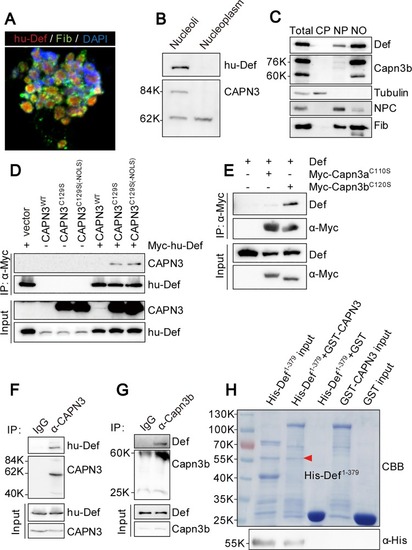
Def directly interacts with CAPN3, and they form a complex in the nucleolus. |
|
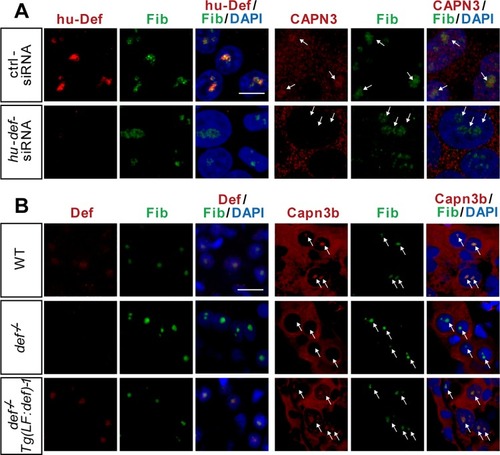
Def determines the nucleolar localisation of Capn3. |
|
Def is phosphorylated at its N-terminus. |
|
S50, S58, S62, S87, and S92 are modified by phosphorylation. |
|
Simultaneous phosphorylations at S58 and S62 or at S87 and S92 are necessary for Def to promote liver development |
|
Simultaneous phosphorylations at S58 and S62 or at S87 and S92 are necessary for Def to promote cell cycle progression. |
|
Def phosphorylation modulates its role in cell cycle in zebrafish and knockdown of CAPN3 or Def in cultured human cells causes a p53-dpendent cell cycle arrest. |
|
Simultaneous phosphorylations at S87 and S92 are necessary for the nucleolar localisation of Capn3b |
|
Simultaneous phosphorylations at S58 and S62 or at S87 and S92 are essential for Def to mediate p53 degradation in the nucleoli. |
|
Def regulates liver development only in part through the p53 pathway. |