- Title
-
The careg element reveals a common regulation of regeneration in the zebrafish myocardium and fin
- Authors
- Pfefferli, C., Jaźwińska, A.
- Source
- Full text @ Nat. Commun.
|
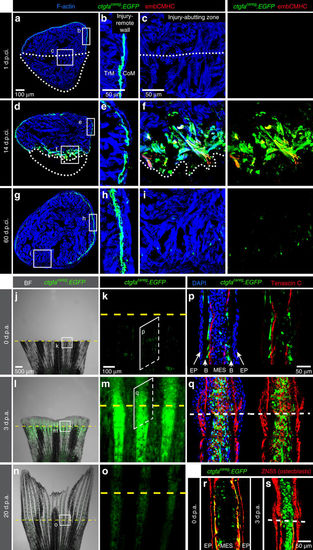
A transgenic reporter of the peri-injured myocardium and the fin stump. (a–i) Transversal ventricle sections of transgenic fish ctgfacareg:EGFP immunostained for embCMHC (red) at different days post-cryoinjury (d.p.ci.). The cardiac muscle is detected by F-actin staining (Phalloidin, blue). (b,e,h) The injury-remote part of the ventricular wall displays a subcortical layer of ctgfacareg:EGFP cells that is located between the thin cortical myocardium (CoM) and the inner trabecular myocardium (TrM). This layer does not express embCMHC and remains unaltered during regeneration. (c,f,i) The injury-abutting zone of the ventricular wall shows transient expression of ctgfacareg:EGFP and embCMHC within a distance of 100 μm from the wound border during regeneration. N≥4. (j–o) Live-imaging of fins at different days post amputation (d.p.a.). Bright-field (BF) was combined with fluorescence. (k,m,o) Higher magnifications of the region at the amputation plane (yellow dashed line) show transient expression of ctgfacareg:EGFP in the stump and the regenerate. N=4. (p–s) Immunofluorescence staining of longitudinal fin sections. (p,r) At 0 d.p.a., ctgfacareg:EGFP+ cells are associated with Tenascin C fibres along the border between the bones (B) and mesenchymal cells (MES), but not the epidermis (EP). Some of these cells express an osteoblast marker visualized with Zns5 antibody (red). (q,s) At 3 d.p.a., Tenascin C demarcates the zone of tissue remodelling in the stump and the outgrowth. ctgfacareg:EGFP is upregulated in the activated MES at the peri-injury zone, but not in Zns5-labelled osteoblasts of the outgrowth. N≥4. Post-infarcted ventricle is encircled with a dotted line. Fin amputation plane is shown with a dashed line. The same rules apply to all subsequent figures. EXPRESSION / LABELING:
|
|
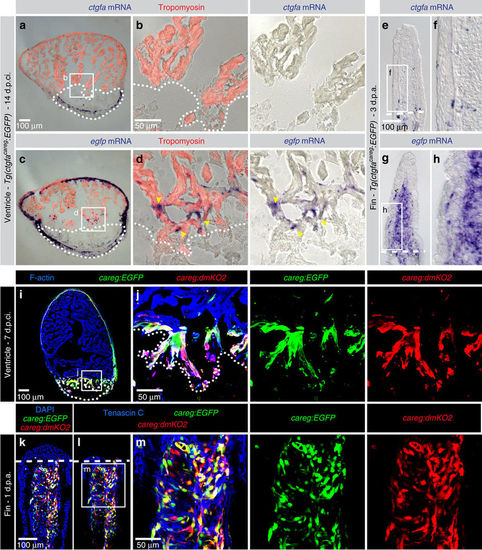
ctgfacareg:EGFP does not reproduce endogenous ctgfa expression in peri-injury tissues. (a–h) In situ hybridization using ctgfa and enhanced green fluorescent protein (egfp) probes on sections of hearts at 14 d.p.ci. and fins at 3 d.p.a. of ctgfacareg:EGFP transgenic fish shows distinct expression patterns. (a,b) ctgfa mRNA is not detected in the myocardium, but in a subset of non-myocytes in the post-infarcted tissue. (c,d) egfp mRNA is expressed in the outer wall of the myocardium and at the peri-injury zone (yellow arrowheads). (e,f) ctgfa mRNA is present in a few bone-associated cells of the fin, while (g,h) egfp mRNA is detected in the blastema. N=6. (i–m) careg:EGFP and careg:dmKO2 double transgenic fish display an overlapping expression pattern in the regenerating heart at 7 d.p.ci. and the fin at 1 d.p.a. N=4. EXPRESSION / LABELING:
|
|
The careg reporter is activated in other injury models of the zebrafish heart and fin. (a,b) Ventricle section of careg:EGFP heart at 7 days post amputation (d.p.a.) labelled with F-actin (blue) and antibodies against GFP (green) and embCMHC (red). careg:EGFP expression is activated in the ventricular trabeculae close to the amputation plane of the resected apex (dashed line). N=3. (c–l) Live-imaging of the same fin at different time points post-cryoinjury. Cryoinjury of the caudal fin results in spontaneous sloughing of destroyed tissue within two days after the damage induction, followed by resumed regeneration. Bright-field was combined with fluorescence. (c,d) At 3 h.p.ci. (hours post-cryoinjury), careg:EGFP is not detected in the fin. Ischaemic tissue, which is distal to the cryoinjury plane (blue line), remains integrated with the rest of the body at this early stage. (e,f) At 1 d.p.ci., the majority of the damaged tissue detached from the stump. careg:EGFP is detected proximally to the damaged zone. (g,h) At 2 d.p.ci., the reparation of the distorted margin is accompanied by careg:EGFP expression. (i,j) At 4 d.p.ci., the protruding blastema displays enhanced expression of the careg reporter. (k,l) The advanced regeneration is associated with downregulation of careg:EGFP expression. N=3. EXPRESSION / LABELING:
|
|
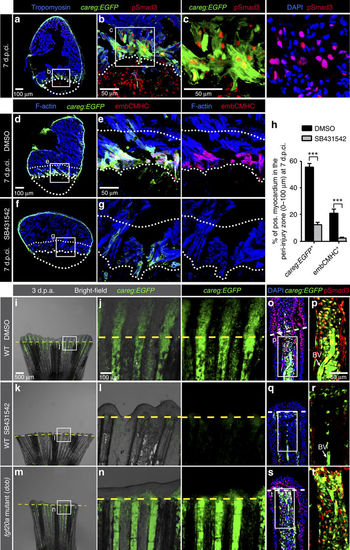
The regeneration biosensor careg is dependent on TGFβ/Activin-β signalling. (a–c) Immunofluorescence staining of careg:EGFP ventricle at 7 d.p.ci. with antibodies against GFP (green), pSmad3 (red) and Tropomyosin (blue) revealed the presence of TGFβ/Activin-β-activated cells in the careg:EGFP-expressing tissue. N=6. (d–g) careg:EGFP heart sections at 7 d.p.ci. treated with 0.1% DMSO or 20 μM SB431542, an inhibitor of TGFβ type I receptors, and immunostained with antibodies against GFP (green) and embCMHC (red). The intact myocardium was detected with F-actin staining (blue). In the magnified images, the upper dotted line demarcates a 100 μm-thick margin of the remaining myocardium from the injury border (lower dotted line). (h) Percentage of careg:EGFP+ and embCMHC+ area within a distance of 100 μm from the post-infarcted tissue in hearts at 7 d.p.ci. treated with DMSO or SB431542. The inhibitor treatment resulted in a significant reduction of careg:EGFP and embCMHC expression in the peri-injured zone, compared to control hearts treated with DMSO. N=8. ***P<0.001; unpaired t-test. Error bars correspond to s.e. of the mean (s.e.m.). (i–n) Live-imaging of careg:EGFP fins at 3 d.p.a. in a wild-type (WT) background treated with DMSO or SB431542, and in the fgf20a (dob) mutant background. careg:EGFP expression is suppressed in the stump by TGFβ inhibition, but it remains normal in fgf20a mutant fins. (o–t) Longitudinal sections of the fins shown in the left panels display a reduction of pSmad3-positive nuclei (red) and careg:EGFP in the stump of SB431542-treated fins, whereas both markers are unaltered in fgf20a mutants. BV, blood vessel. N=4. |
|
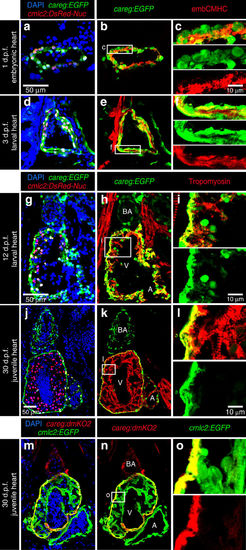
careg is expressed in embryonic CMs and the outer wall of developing ventricle. (a–l) Longitudinal sections of careg:EGFP;cmlc2:DsRed2-Nuc double transgenic hearts at different time points during development. Cardiac nuclei are marked by DsRed expression. The endogenous fluorescence was quenched with HCl treatment before immunostaining. GFP and DsRed were detected by antibody staining. (a–f) At 1 and 3 d.p.f. (days post-fertilization), careg:EGFP and endogenous embCMHC are co-expressed in embryonic CMs. (g–i) At 12 d.p.f., a few careg:EGFP+ CMs delaminate from the outer heart surface and invade into the ventricle (V) chamber, as seen by the residual EGFP. (j–l) At 30 d.p.f., careg:EGFP is restricted to the outer layer of the ventricular wall, and is downregulated in the trabecular myocardium. careg:EGFP is also expressed in non-myocytes of the bulbus arteriosus (BA) and a few cells of the atrium (A). (m–o) Longitudinal sections of careg:dmKO2;cmlc2:EGFP transgenic heart at 30 d.p.f. immunostained for GFP (green) and dmKO2 (red). The careg:dmKO2 transgenic line has a cardiac developmental expression pattern similar to that of careg:EGFP (Supplementary Fig. 10). N≥5. |
|
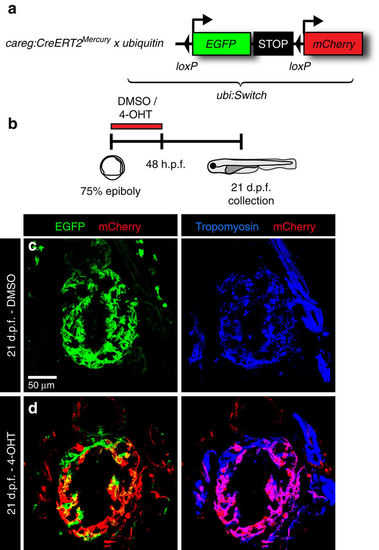
Embryonic careg-positive CMs contribute to the trabecular myocardium. (a) Schematic representation of the transgenic strains used for lineage tracing. (b) Experimental design. (c,d) Longitudinal sections of hearts at 21 d.p.f. immunostained against mCherry (red), GFP (green) and Tropomyosin (blue). (c) Control embryos treated with the vehicle do not display mCherry fluorescence. (d) mCherry+ CMs are present in the trabecular and outer myocardium at 21 d.p.f. in embryos treated with 4-OHT, suggesting that the trabecular myocardium derives from embryonic careg+ CMs. N=9. |
|
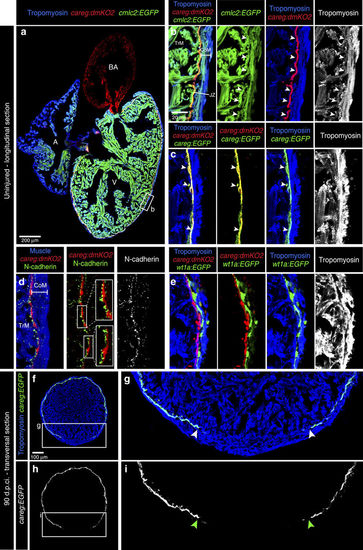
Monitoring the primordial CM layer between the compact and trabecular myocardium. (a) Longitudinal sections of careg:dmKO2;cmlc2:EGFP transgenic adult hearts labelled with antibodies against KO2 (red) and Tropomyosin (blue, white). V, ventricle; A, atrium; BA, bulbus arteriosus. (b) careg:dmKO2 expression is maintained during adulthood in a single layer of thin subcortical CMs (arrowheads) in the junctional zone (JZ) between the compact/cortical (CoM) and the trabecular myocardium (TrM). N=4. (c) A higher magnification of the adult ventricle wall of double transgenic fish careg:dmKO2; careg:EGFP displays an overlapping expression of both markers in subcortical myocytes (arrowheads). N=4. (d) A magnified adult ventricle wall of careg:dmKO2 transgenic fish displays an abundant expression of N-cadherin (green, white) on the surface of the junctional CMs on the side facing the trabecular myocardium. N=4. (e) A fragment of the adult ventricle wall of careg:dmKO2;wt1a-6.8kb:GFP transgenic fish displays the separation of compact myocardium and subcortical careg:dmKO2+ layer by wt1a-6.8kb:GFP-labelled fibroblasts of the junctional zone. N=4. (f–i) At 90 d.p.ci., transversal heart sections of careg:EGFP hearts display a gap in the primordial layer within the regenerated myocardium. The extent of the gap is indicated with arrowheads. N=4. |
|
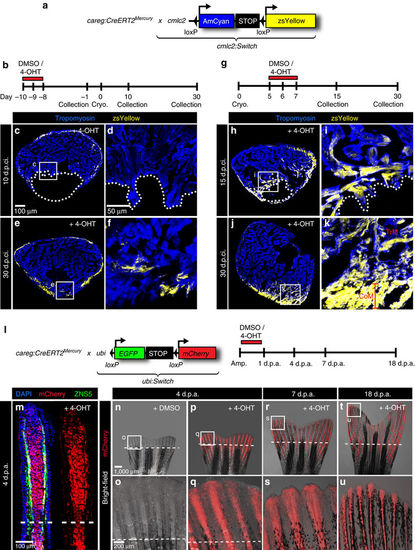
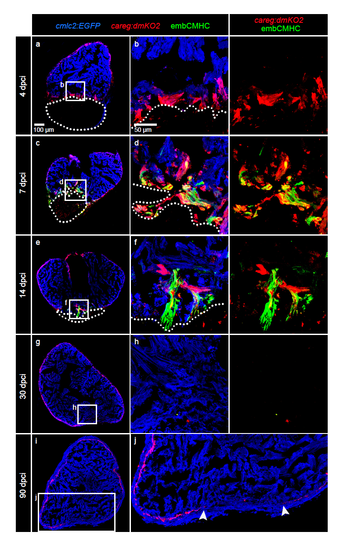
careg-expressing cells contribute to the regenerating myocardium and fin mesenchyme. (a,b) Schematic representation of the transgenic strains and the experimental design for lineage tracing of careg-expressing primordial CMs during ventricle regeneration. (c–f) Representative sections of hearts treated with 4-OHT before cryoinjury to label the primordial layer, stained with antibody against Tropomyosin (blue). (c,d) At 10 d.p.ci., no zsYellow is detected in the trabecular myocardium at the injury site, indicating that primordial CMs do not contribute to the regeneration zone along the post-cryolesioned tissue. (e,f) At 30 d.p.ci., the zsYellow-positive primordial layer regenerates incompletely. (g) Experimental design for lineage tracing of careg-expressing dedifferentiated CMs during ventricle regeneration. (h–k) Representative sections of hearts treated with 4-OHT after cryoinjury hearts immunostained against Tropomyosin (blue) (h,i) At 15 d.p.ci., zsYellow is expressed in the trabecular myocardium at the injury site. (j,k). At 30 d.p.ci., zsYellow-positive CMs are detected in the trabecular (TrM) and cortical myocardium (CoM) of the regenerated muscle. N≥8. (l) Transgenic strains and the design of lineage tracing experiments during fin regeneration. (m) Longitudinal section of lineage-labelled fins at 3 d.p.a., stained for osteoblasts using the Zns5 antibody (green) and for nuclei with DAPI (blue). mCherry is detected in the entire mesenchyme of the regenerative outgrowth and below the amputation plane. N=6. (n–u) Live-imaging of the same lineage-labelled fin at different time points of regeneration. The regenerative outgrowth and the stump tissue are labelled with mCherry. N=6. |
|
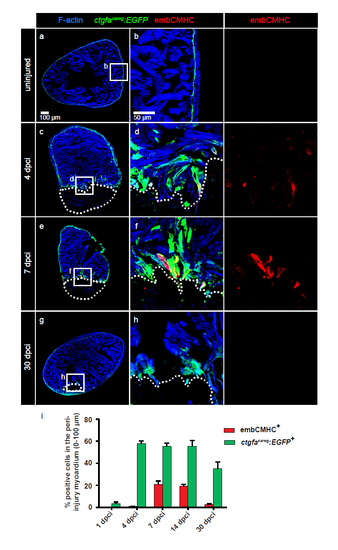
The ctgfacareg:EGFP transgenic reporter is transiently activated in the peri-injured myocardium during heart regeneration. (a-h) Immunofluorescence staining of ctgfacareg:EGFP transversal heart sections with antibodies against GFP (green) and embCMHC (red) at different time points after cryoinjury. The intact myocardium is detected by F-actin staining (Phalloidin, blue). (a-b) In uninjured ventricle, ctgfacareg:EGFP is expressed in a fine layer in the subcortical myocardium. No embCMHC is observed. (c-d) At 4 dpci, ctgfacareg:EGFP is induced in trabecular fascicles abutting the post-infarcted area, a subset of which also displays embCMHC expression. (e-f) At 7 dpci, ctgfacareg:EGFP expression is maintained at the injury border covering embCMHC+ CMs. (g-h) At 30 dpci, ctgfacareg:EGFP is reduced to a small margin around the remaining fibrotic tissue. (i) Quantification of ctgfacareg:EGFP+ and embCMHC+ area within 100 μm from the injury border. N ≥ 8. Post-infarcted ventricle is encircled with a dotted line. The same rules apply to all subsequent figures. |
|
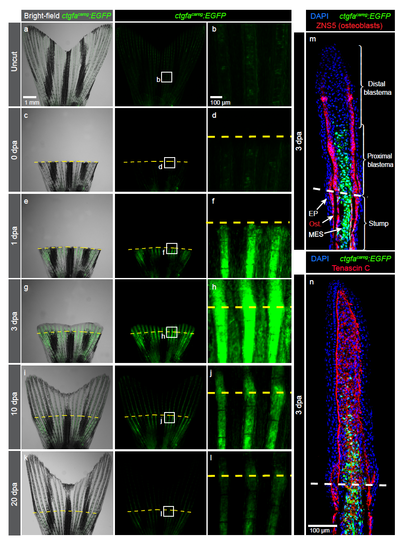
Peri-injury tissue of the fin stump transiently induces ctgfacareg:EGFP expression (a-l) Live imaging of ctgfacareg:EGFP fins at different days post-amputation (dpa). Higher magnifications of framed areas show the region of the amputation plane (yellow dashed line). N=4. (m-n) Immunofluorescence staining of longitudinal ctgfacareg:EGFP fin sections at 3 dpa. EP, epidermis; Ost, osteoblasts; MES, mesenchmyme. (m) ctgfacareg:EGFP is detected in the stump mesenchyme and osteoblasts (red) below the amputation plane (dashed line), and in the proximal blastema. (n) The mesenchyme of the regenerating fin abundantly expresses Tenascin C, a tissue remodelling extracellular protein. N≥4. |
|
careg:dmKO2 is transiently expressed in the peri-injured myocardium during heart regeneration. (a-j) Immunofluorescence staining of careg:dmKO2;cmlc2:EGFP heart sections labelled with antibodies against GFP (cardiac cells, blue) and embCMHC (green) at different time points after cryoinjury. N=4. (a-b) At 4 dpci, careg:dmKO2+ CMs emerge along the wound margin. (c-f) At 7 and 14 dpci, a large proportion of careg:dmKO2+ CMs activates the expression of embCMHC. (g-h) At 30 dpci, careg:dmKO2 expression declines in the trabecular myocardium, but the primordial layer was not fully restored in the regenerated part of the ventricle. (i-j) At 90 dpci, the myocardium is completely regenerated, but the expression of careg:dmKO2 is not completely reestablished in the subcortical region. The arrowheads indicate the position of the gap in the subcorical layer. |
|
careg:dmKO2 is transiently expressed in the peri-injured stump during fin regeneration. (a-l) Live-imaging of careg:dmKO2 fins at different days post-amputation (dpa). Higher magnifications of framed areas show the region of the ampulation plane (yellow dashed line). N=4. |
|
The careg reporter is activated in other injury models of tissue regeneration in zebrafish. (a-d) In-situ hybridization (purple) on longitudinal fin sections reveals that, as in the amputation model, the endogenous ctgfa gene is not upregulated in the regenerating fin after cryoinjury. (a, c) A probe against a blastema marker, msxb, was used as a positive control to detect the activated mesenchyme. (b, d) A ctgfa probe does not label the regenerating fin tissues. M, mesenchyme; E, epidermis. Brackets indicate the damaged tissue remaining at the tip of the cryoinjured stump. N=3. (e-i) Live-imaging of the larval tail of careg:dmKO2 fish at different days post-fertilization (dpf) and post-amputation. The transgenic reporter is constitutively expressed in the notochord. After amputation (dashed line), the expression of careg:dmKO2 is markedly upregulated at the tip of the regenerating tail. N=6. |
|
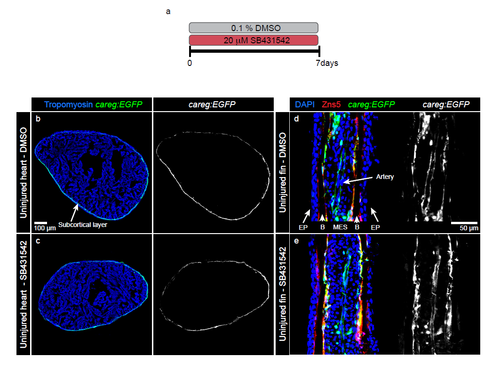
Treatment of uninjured organs with the inhibitor of TGFβ/Activin-β signalling does not affect the homeostatic expression of careg:EGFP. (a) Experimental design. Uninjured fish were treated with the inhibitor of TGFβ/Activin-β (20 μM SB431542) for 7 days. (b-c) Immunofluorescence staining of uninjured ventricles after 7 days of SB431542 treatment. The expression of careg:EGFP in the subcortical layer is similar in control and treated fish. (d-e) Longitudinal sections of rays in uninjured fins. Osteoblasts are marked with Zns5 antibody (red). They cover the surface of acellular bone matrix. In both control and treated specimens, careg:EGFP is expressed in intraray osteoblasts, in a few scattered MES and the artery. EP, epidermis; MES, mesenchyme; B, accelular bones. N=4. |
|
The inhibition of TGFβ/Activin-β signaling suppresses CM dedifferentiation and proliferation at the injury-abutting zone during heart regeneration. (a-d, f-i) Immunofluorescence staining of ventricles at 7 dpci (a-d) or 14 dpci (f-i) treated with DMSO or 20 μM SB431542 labelled with antibodies against fluorescent proteins and embCMHC. The intact myocardium is detected by F-actin staining (blue). The upper dotted lines indicate the 100 μm-thick margin of the myocardium along the injury border. (e, j) Percentage of positive myocardium for the careg reporter or embCMHC in a 100 μm-wide margin of the regenerating myocardium in control or SB431542-treated hearts. N ≥ 5. ***p < 0.0001, **p < 0.01, *p < 0.01; unpaired t-test. Error bars correspond to standard error of the mean (SEM). (k-n) Immunofluorescence staining of careg:EGFP ventricle at 7 dpci treated with DMSO or 20 μM SB431542 labelled with antibodies against GFP (blue), Mef2c (cardiac nuclei, red) and BrdU (green). (o) Percentage of BrdU+ CMs in a 100 μm-wide margin of the regenerating myocardium in control or SB431542-treated hearts. N ≥ 5. ***p < 0.0001; unpaired t-test. Error bars correspond to standard error of the mean (SEM). |
|
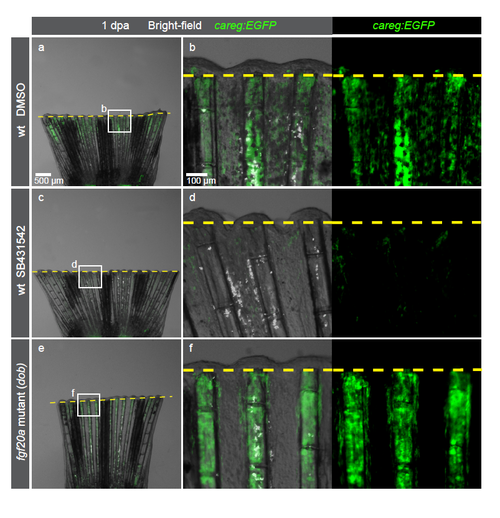
Despite an absence of the regenerative outgrowth, dob mutant fins normally induce careg:EGFP in the amputation-activated stump. (a-f) Live-imaging of careg:EGFP fins at 1 dpa in a wild-type (wt) background treated with DMSO or SB431542, and in the fgf20a (dob) mutant background. careg:EGFP expression is suppressed in the stump by the inhibition of TGFβ/Activin-β signaling, but it remains normal in fgf20a mutant fins. N=4. |
|
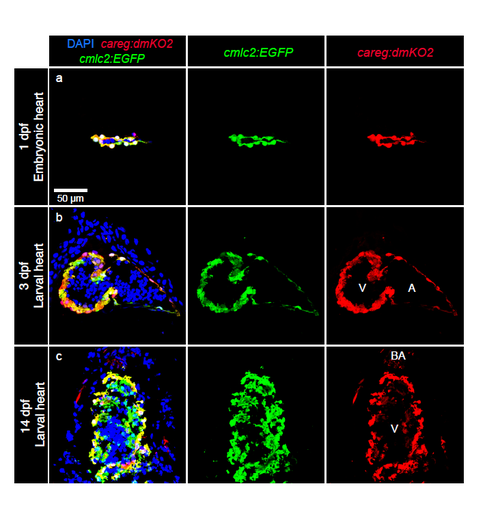
Developmental dynamics of careg:dmKO2 expression in the zebrafish heart. (a-c) Longitudinal sections of careg:dmKO2;cmlc2:EGFP transgenic hearts at different time points during development, labelled with DAPI (nuclei, blue) and antibodies against GFP (green, cardiac cells) and KO2 (red). (a, b) careg:dmKO2 is expressed in all embryonic cardiomyocytes at 1 and 3 dpf. (c) At 14 dpf, careg:dmKO2 is maintained in the outer compact layer of the ventricle, whereas it is downregulated in the trabecular myocardium. careg:dmKO2 is also expressed in the bulbus arteriosus. N ≥ 5. dpf, days post-fertilization; V, ventricle; BA, bulbus arteriosus; A, atrium. |
|
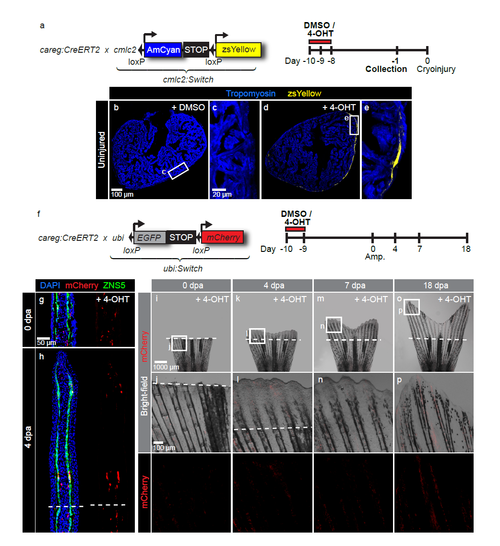
Control experiments for careg lineage tracing analysis of heart and fin regeneration. (a) Schematic representation of the transgenic strains and the experimental design for lineage tracing of careg-expressing primordial CMs during ventricle regeneration. (b-e) Immunostaining of uninjured ventricles treated with DMSO or 4-OHT using Tropomyosin (blue). Exposure to 4-OHT for 2 days is sufficient to label primordial CMs with zsYellow, as analysed at 7 days after the treatment (-1 day). (f) Schematic representation of the transgenic strains and the experimental design for lineage tracing of careg-expressing cells during fin regeneration. (g, h) Longitudinal fin sections at 0 and 4 dpa immunostaining for mCherry (red) and Zns5 (green). Exposure to 4-OHT for 1 day results in labelling of a few osteoblasts. (i-p) Live imaging of fins at different dpa after treatment with 4-OHT reveals only a few mCherry labelled cells in the stump and the regenerate. N ≥ 4. |