- Title
-
MiR-10 Represses HoxB1a and HoxB3a in Zebrafish
- Authors
- Woltering, J.M., and Durston, A.J.
- Source
- Full text @ PLoS One
|
Spatial and temporal expression profile of miR-10c. A) RT-PCR with primers located 5′ of miR-10c and within the coding region of exon 1 of HoxB4a shows inclusion of miR-10c and HoxB4a on the same transcript. PCR 35 cycles, -RT: no reverse transcriptase added. B) RT-PCR shows similar temporal expression during development of HoxB4a (28 cycles) and miR-10c pre-miRNA (35 cycles). C) Whole mount in situ hybridization on different stage Zebrafish embryos shows mutually exclusive expression of the HoxB3a rhombomere 5/6 domain (red) with miR-10c (purple). EXPRESSION / LABELING:
|
|
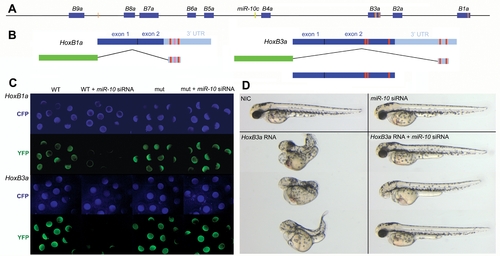
miR-10 target sites within the HoxBa cluster. A) Schematic representation of the zebrafish HoxBa cluster with MiR-10 seed sequences (nucleotide 1-7) within the sense strand indicated as orange bars. Known and EST database inferred mature Hox transcripts are indicated in blue. The miR-10c microRNA gene is indicated in green. B) Schematic representation of the HoxB1a and HoxB3a E-YFP sensor constructs and the HoxB3a overexpression construct. Red boxes indicate the position of the seed sequences. The light red box in the HoxB1a 3′ UTR is a target site flanked at position one by a T instead of an A. C) Validation of the HoxB1a and HoxB3a E-YFP sensor constructs by injection of wildtype (WT) and seed mutant (mut) constructs in presence and absence of miR-10 siRNA. E-CFP was co-injected as a loading control. D) Phenotypic sensor assay to validate the HoxB3a ORF miR-10 target sites. Overexpression of 40pg HoxB3a results in severe anterior and posterior truncations that are rescued by co-injection with miR-10 siRNA |
|
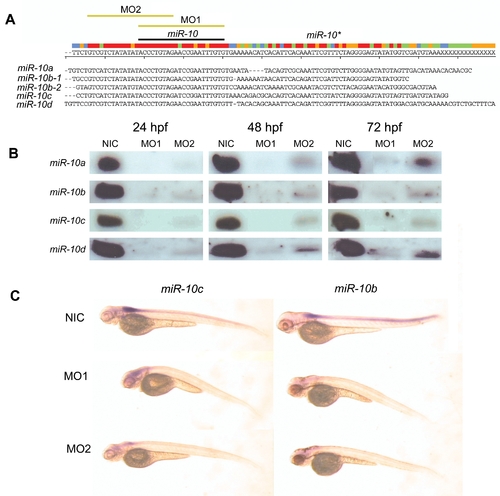
Morpholino knockdown of miR-10 A) ClustalW alignment of the 5 Zebrafish miR-10 precursor sequences. Indicated are the positions of the mature microRNA, the hairloop and the miR-10* (antisense pairing sequence in the hairpin). The target sequences for both morpholino reagent 1 and 2 are indicated with yellow bars (MO1 and MO2). B) Northern blot for all 4 different miR-10 isoforms in morpholino injected embryos at 24, 48 and 72 hpf. There is an absence or very strong downregulation of the mature microRNA in the morpholino injected samples. C) LNA in situ hybridization for miR-10b and miR-10c in 72 hpf miR-10 morphants. The endogenous expression of miR-10b and miR-10c is no longer detected. EXPRESSION / LABELING:
|
|
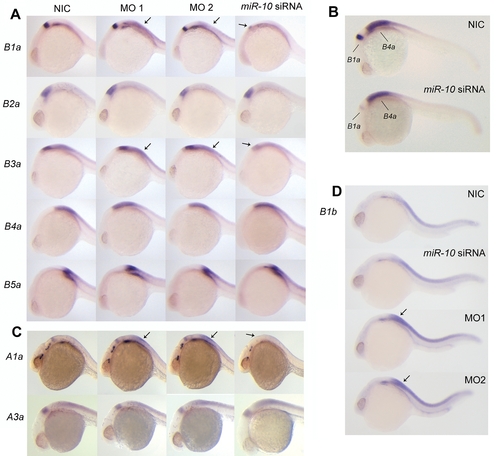
Effect of miR-10 knockdown and overexpression of endogenous Hox target transcripts A) Whole mount in situ hybridization with probes for hoxB1a, B2a, B3a, B4a and B5a on 24hpf embryos injected with morpholino reagent 1 or 2 (MO1 or MO2), miR-10 siRNA or non injected controls (NIC). To allow quantitative detection, embryos hybridized with the same probe were stained equally long and staining was continuously monitored and stopped before reaching signal saturation. HoxB1a and HoxB3a respond to both gain and loss of function (arrows) of miR-10 with a decrease and increase in expression levels respectively. HoxB2a, HoxB4a and HoxB5a are unresponsive to miR-10 overexpression or knockdown. B) Double whole mount in situ hybridization on 24 hr embryos using probes for hoxB1a and hoxB4a showing the different responses of the genes. Embryos were stained equally long till adequate staining was obtained for the hoxB4a probe. C) In situ hybridization with HoxA1a and HoxA3a on 24 hpf embryos injected with MO1, MO2 or miR-10 siRNA. HoxA1a responds to both overexpression and knockdown. The transcript level of HoxA3a does not respond to either overexpression or knockdown. D) In situ hybridization with HoxB1b on 24 hpf embryos morphant and overexpression embryos. There is strong upregulation of HoxB1b expression in the morphants but no downregulation is observed in the miR-10 siRNA injected embryos. EXPRESSION / LABELING:
|
|
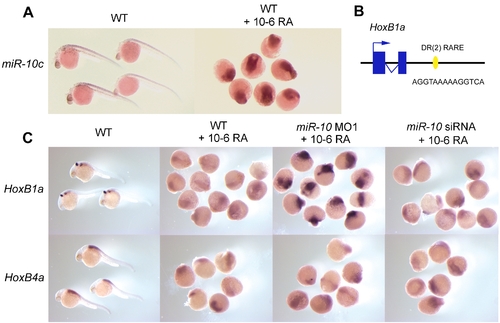
Retinoid induction of miR-c10 and upregulation of HoxB1a in miR-10. morphants. A) LNA in situ hybridization for miR-10c in wildtype (WT) and 10-6M retinoic acid (RA) treated embryos. RA treatement results in miR-10c upregulation. B) Presence of a DR[2] type retinoic acid response element (RARE) 1kb 3′ of the Zebrafish HoxB1a gene. This sequence is conserved in the mouse in which it has been shown to mediate the neural response of HoxB1 to RA [38] C) Different response of HoxB1a to RA stimulation in wildtype or miR-10 morphant embryos. HoxB1a is strongly upregulated in miR-10 morphants. Injection with the miR-10 siRNA has no effect. HoxB4a responds similar to all conditions. EXPRESSION / LABELING:
|
|
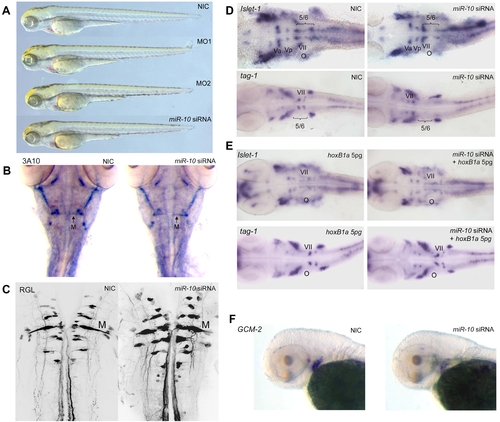
Overexpression of miR-10 induces HoxB1a and HoxB3a loss of function phenotypes. A) Wildtype (WT), miR-10 morphant (MO1, MO2) and miR-10 siRNA overexpression embryos at 72 hpf show no apparent developmental differences. B) Mauthner neuron development as visualized by 3A10 neurofilament immunostaining in 72 hpf embryos shows no differences between miR-10 siRNA injected embryos and controls. C) Confocal images of reticulospinal hindbrain neurons in retrograde labeled, 5 day old embryos. Wildtype and miR-10 siRNA injected embryos are similar. D) Islet-1 and tag-1 in situ hybridization on 30 hpf wildtype and miR-10 siRNA injected embryos. Flatmounts of head regions are shown. In wildtype embryos the VIIth cranial nerve migrates into rhombomere 5/6 at the level of the otic vesicle. In miR-10 siRNA injected embryos the VIIth nerve does no longer migrate out of rhombomere 4. E) Co-injection of 5pg HoxB1a RNA rescues the miR-10 siRNA induced migration defect of the VIIth cranial nerve as shown by islet-1 and tag-1 in situ hybridization. F) Gcm-2 expression is downregulated in miR-10 siRNA injected embryos, which is consistent with repression of HoxB3a. EXPRESSION / LABELING:
|
|
MiR-10 acts in synergy with HoxB4 A) Embryos injected with HoxB4, miR-10 siRNA and HoxB4+miR-10 siRNA analyzed for the expression of HoxB1a and HoxB3a at 24 hpf. Injection of 150pg HoxB4 leads to downregulation of HoxB1a and of downregulation of the hindbrain domain of HoxB3a. The rhombomere 4 expression domain of HoxB1a and the rhombomere 5/6 expression domain of HoxB3a are still discernable though. When 150pg HoxB4 is expressed together with miR-10 siRNA the expression domain of HoxB1a disappears and no clear rhombomere 5/6 stripe of HoxB3a expression can be detected. B) Embryos injected with HoxB4, miR-10 siRNA and HoxB4+miR-10 siRNA analyzed for the expression of endogenous HoxB4a at 48 hpf. The combination of HoxB4 together with the miR-10 siRNA induces a stronger phenotype with more severe anterior and posterior truncations than injection with HoxB4 alone. On the right groups of embryos injected with HoxB4 or the combination of HoxB4 and miR-10 siRNA are shown. |
|
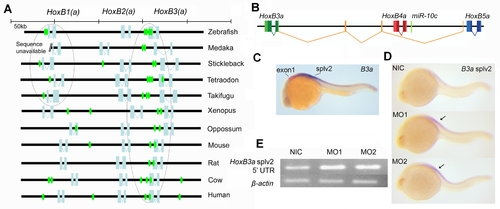
Evolutionary conservation of miR-10 targetsites and autoregulation of miR-10c A) Putative miR-10 target sites are indicated by seed sequences in the sense strand of the anterior vertebrate HoxB(a) clusters. Seed sequences are shown in green, open reading frames are indicated in light blue. Note conserved association of target sites with the HoxB3(a) ORF and conserved presence of a putative target site in Teleost HoxB1a. B) The HoxB3a splv2 polycistronic transcript includes one exon between HoxB4a and HoxB5a, two exons between HoxB4a and HoxB3a and the main HoxB3a coding sequence. The primary transcript for this isoforms includes miR-10c. The 52 UTR sequence is shown in orange, this sequence corresponds to the probe used in C and D to specifically detect this splice isoforms. C) Comparison of the HoxB3a exon1 expression (red) and the expression of HoxB3a splv2 (purple). HoxB3a splv2 is expressed posterior to the main rhombomere 5/6 expression domain of HoxB3a as reported previously [46]. The staining reaction for HoxB3a splv2 was developed for much longer than the reaction for the HoxB3a exon1 probe and the HoxB3a splv2 is presumably expressed at a much lower level. D) In situ hybridization with HoxB3a splv2. Expression is upregulated in miR-10 morphant embryos (arrows). E) Semi quantitative RT-PCR for the HoxB3a splv2 52 UTR, ß-actin is used as loading control. HoxB3a is upregulated in miR-10 morphant embryos. HoxB3a splv2: 31 cycles, ß-actin: 22 cycles. |
|
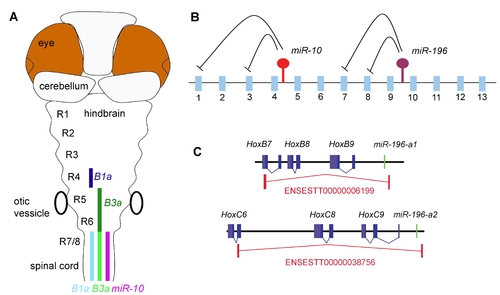
Post-transcriptional regulatory interactions with the hox clusters. A) Schematic representation of miR-10 and target gene expression in the Zebrafish hindbrain. MiR-10 is expressed posterior from the rhombomere 6/7 boundary. The target genes HoxB1a and HoxB3a are expressed in a strong domain (dark colour) anterior in the anterior hindbrain and in a weaker domain (light colour) in the area where they overlap with miR-10. HoxB1a shows a gap in expression in r5 and 6, possibly due to stronger transcriptional repression. B) Schematic representation of the post-transcriptional relations within the hox clusters. MiR-196 is known to represses HoxB8, HoxC8, HoxD8 and HoxA7 and we have identified HoxB1a and HoxB3a as targets for miR-10. The emerging view is that the microRNAs in the hox clusters target more anterior genes in their close proximity. C) Polycistronic transcripts identified from the EST database show inclusion of miR-196 paralogues and HoxB8 and HoxC8 target genes on the same primary transcripts. |
|
Expression of HoxB3a exon1 and HoxB3a sensor regions EXPRESSION / LABELING:
|
|
Double in situ hybridization of HoxB3a with miR-10a, miR-10b, miR-10c and miR-10d EXPRESSION / LABELING:
|
|
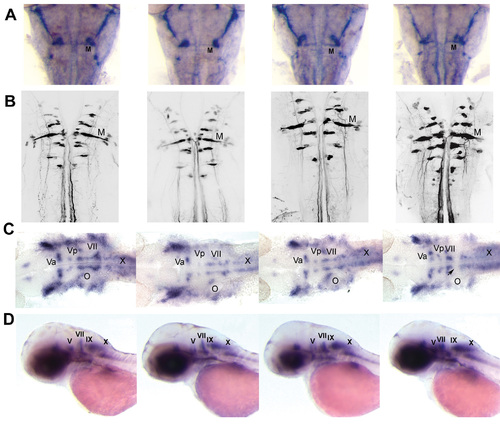
Patterns of primary and secondary hindbrain motorneurons in miR-10 overexpression and morphant embryos |