- Title
-
Zebrafish Dapper1 and Dapper2 play distinct roles in Wnt-mediated developmental processes
- Authors
- Waxman, J.S., Hocking, A.M., Stoick, C.L., and Moon, R.T.
- Source
- Full text @ Development
|
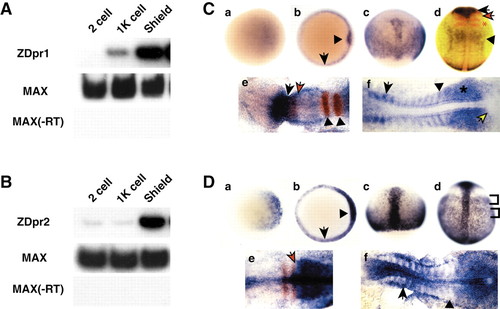
RT-PCR and in situ hybridization analysis of zebrafish dpr1 and dpr2. (A,B) RT-PCR analysis (25 embryos per stage) of dpr1 (A) and dpr2 (B). Zebrafish max is used as a loading/positive control. –RT is a control for genomic DNA contamination. (C,D) In situ hybridization for expression of dpr1 (C, parts a-f) and dpr2 (D, parts a-f), with embryos shown that are representative of at least 25 analyzed per stage. Stages: (a) sphere stage; (b) shield stage; (c) 80% epiboly; (d) tailbud; (e) five somites (in C), three somites (in D); (f) 12 somites. Orientation: (a,b) animal views, dorsal is rightwards; (c,d) dorsal views, anterior is upwards; (e,f) anterior is leftwards. (C, part b) Arrow indicates margin and arrowhead indicates shield. (C, part d) dpr1 (blue), pax2.1 (red, midbrain/hindbrain boundary) and krox20 (red, –r3 and 5). Black arrow indicates dpr1 expression in brain. Black arrowhead indicates dpr1 expression in anterior spinal cord. Red arrow indicates pax2.1. Red asterisk indicates krox20. (C, part e) Black arrow is posterior limit of dpr1(blue) expression. Red arrow is posterior limit of pax6.1(red) expression. Arrowheads indicate r3 and r5. (C, part f) Arrow indicates anterior older somite. Arrowhead indicates staining of posterior, younger somite. Asterisk indicates presomitic mesoderm. Yellow arrow indicates tailbud. (D, part b) Arrow indicates margin and arrowhead indicates shield. (D, part d) Brackets indicate mediolateral banding pattern of staining in lateral mesoderm. (D, part e) Arrow indicates boundary of krox20 (red) and dpr2 (blue) staining. (D, part f) Arrow indicates staining in the anterior of the older somites. Arrowhead indicates presomitic mesoderm. EXPRESSION / LABELING:
|
|
Zebrafish dpr1 and dpr2 morphant phenotypes. (A) RT-PCR analysis of efficacy of dpr1 MO. dpr1 splice blocking MO induces the predicted shift to an 1700 band, indicating it abrogates proper splicing of the transcript. Spliced band is at 500 bp. U, uninjected; I, injected. (B) 5'-Dpr2-luciferase construct is blocked in an in vitro transcription/translation reaction. ß-Galactosidase was used as an internal control. (C) dpr1 morphants (b, injected with 12 ng MO) are discernibly smaller but have no major change of cell fates compared with control (a). dpr2 morphants (c, injected with 8 ng MO) have convergent extension defects, which are more apparent in whole embryos (D, parts c,d). However, the notochord is noticeably wider – compare distances between the small posterior arrowheads in c with a and b. dpr1+dpr2 morphants (d) do not have novel phenotypes, indicating they are not redundant. White arrow indicates anterior limit of head. Black arrows indicate distance between opl and en2. Small arrowhead indicates somite. Posterior arrowheads indicate width of notochord. Embryos were flatmounted, with anterior towards the left. (D) Whole-mount in situ hybridizations of dpr2 morphants indicate they are shorter (compare arrowheads in a and c) and the somites and notochord are wider (compare myod staining in b and d), but major specification events are not affected. For C and D, the experiments were repeated four times with comparable results, see text for penetrance of phenotypes. In situ probes used from anterior to posterior were: cathepsinL (catL-hatching gland), opl (telencephalon), en2 (midbrain/hindbrain boundary), krox20 (rhombomeres 3 and 5) and myod (adaxial cells and somites). PHENOTYPE:
|
|
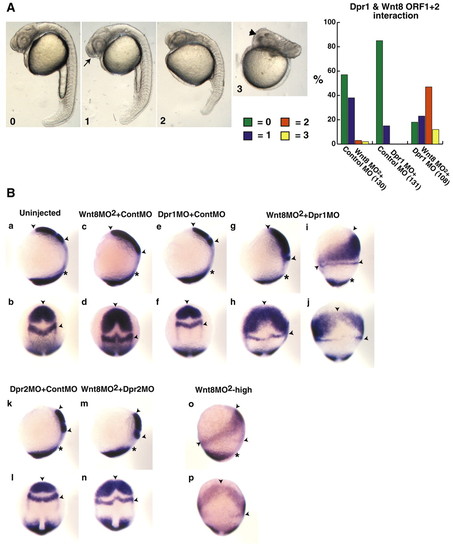
Dpr1 is an enhancer of Wnt/ß-catenin signaling required for ventral and posterior cell fates. (A) Representative embryos for scoring of ventroposteriorization at 24 hours. 0, wild type; 1, slight enlargement of the telencephalon (arrow); 2, strong anteriorization phenotype, enlargement of the head and reduction of the tail; and 3, very strong anteriorization phenotype, super enlargement of the telencephalon (arrowhead) and major loss of trunk and tail. Graph shows percentage of embryos with the respective phenotypes (numbers in brackets indicate the number of embryos used). (B) dpr1, dpr2 and wnt8MO2 morphant phenotypes at the one- to two-somite stage. In situ markers used were opl (telencephalon), pax2.1 (midbrain/hindbrain boundary) and tbx6 (ventrolateral mesoderm). Arrowheads indicate distance between anterior limit of opl and posterior limit of pax2.1. Asterisks indicate anterior limit of tbx6. (a,c,e,g,i,k,m,o) Dorsal is towards the right. (b,d,f,h,j,l,n,p) Dorsal is out of the plane of the page. In all figures, anterior is upwards. Suboptimal dose of wnt8 MO2 is 0.45 ng each MO (0.9 ng total). dpr1 MO dose is 12 ng. dpr2 MO dose is 8 ng. The experiments were repeated five times with comparable results; see text for penetrance of phenotypes. PHENOTYPE:
|
|
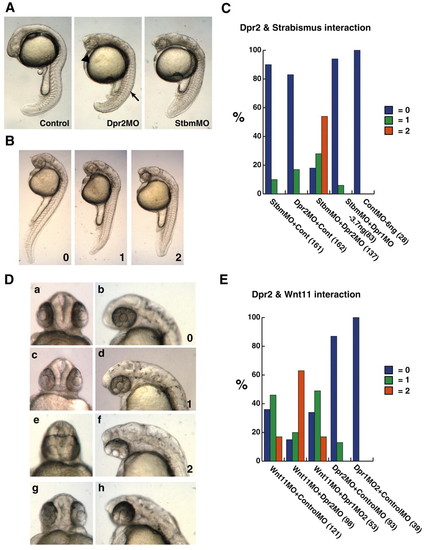
dpr2 morphants phenocopy strabismus/trilobite (stbm) morphants, and functionally interact with stbm and wnt11/silberblick. (A) dpr2 morphants (middle-injected with 7.5 ng dpr2 MO) phenocopy stbm morphants (right-injected with 3 ng). Arrow indicates flattened, non-chevron-shaped somite. Arrowhead indicates narrowed eyes. (B) Scoring of embryos that were co-injected with suboptimal doses of stbm and dpr2 MOs. 0, wild type; 1, moderate CE defects; 2, severe CE defects. (C) Percentage of embryos injected with MOs exhibiting phenotype. (D) Scoring of cyclopia index of embryos co-injected with dpr2 MO and wnt11 MO (a-f). dpr2 MO (normal dose, g,h). 0, wild type; 1, narrowing of the eyes; 2, fusion of the lens. (E) Percentage of embryos injected displaying respective phenotypes. Suboptimal (hypomorphic) dose of dpr2 MO is 3 ng, stbm MO is 0.4 ng and wnt11 MO is 1.5 ng. All injections were balanced with control MO. The numbers in brackets indicate the numbers of embryos used. PHENOTYPE:
|