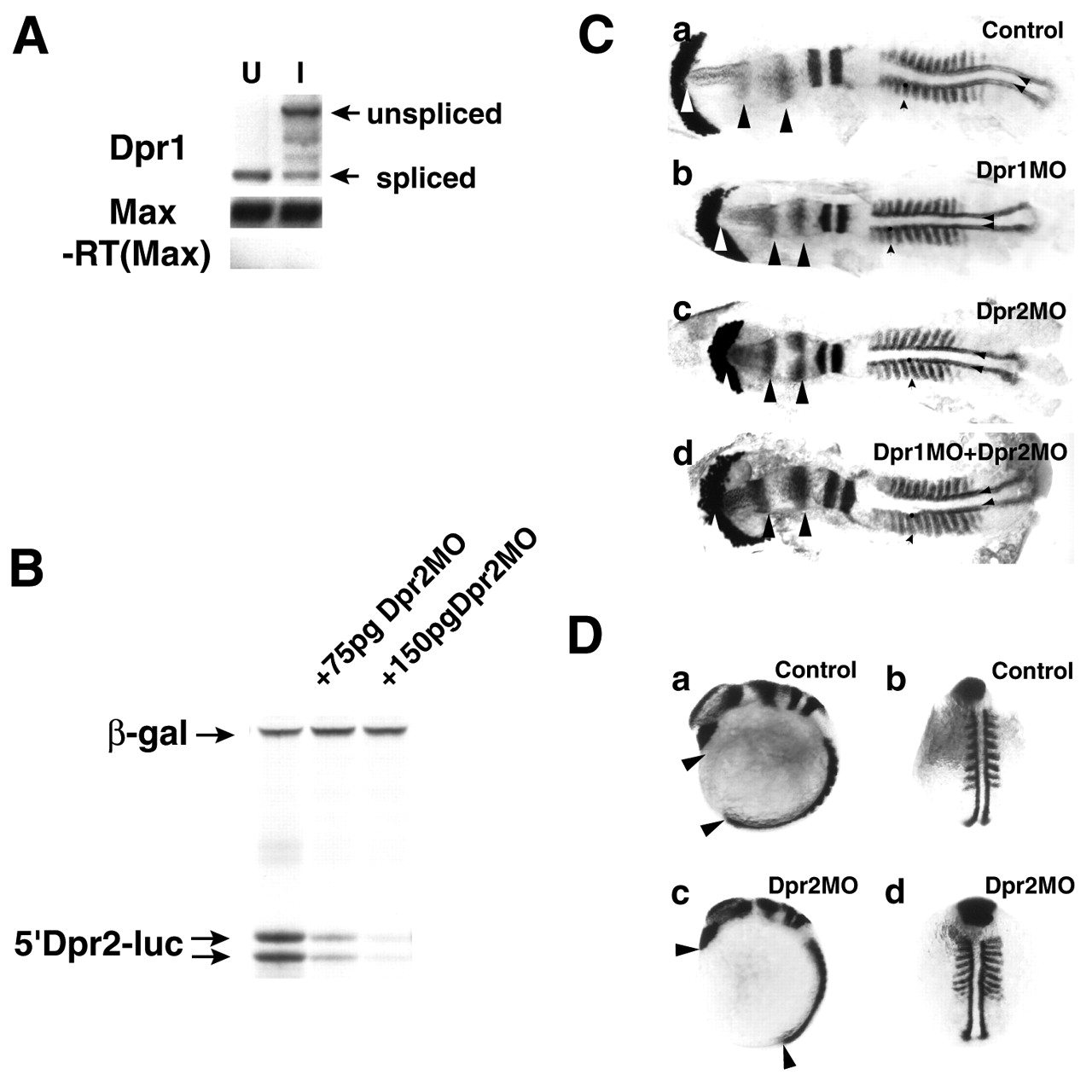
Fig. 3 Zebrafish dpr1 and dpr2 morphant phenotypes. (A) RT-PCR analysis of efficacy of dpr1 MO. dpr1 splice blocking MO induces the predicted shift to an 1700 band, indicating it abrogates proper splicing of the transcript. Spliced band is at 500 bp. U, uninjected; I, injected. (B) 5'-Dpr2-luciferase construct is blocked in an in vitro transcription/translation reaction. ß-Galactosidase was used as an internal control. (C) dpr1 morphants (b, injected with 12 ng MO) are discernibly smaller but have no major change of cell fates compared with control (a). dpr2 morphants (c, injected with 8 ng MO) have convergent extension defects, which are more apparent in whole embryos (D, parts c,d). However, the notochord is noticeably wider – compare distances between the small posterior arrowheads in c with a and b. dpr1+dpr2 morphants (d) do not have novel phenotypes, indicating they are not redundant. White arrow indicates anterior limit of head. Black arrows indicate distance between opl and en2. Small arrowhead indicates somite. Posterior arrowheads indicate width of notochord. Embryos were flatmounted, with anterior towards the left. (D) Whole-mount in situ hybridizations of dpr2 morphants indicate they are shorter (compare arrowheads in a and c) and the somites and notochord are wider (compare myod staining in b and d), but major specification events are not affected. For C and D, the experiments were repeated four times with comparable results, see text for penetrance of phenotypes. In situ probes used from anterior to posterior were: cathepsinL (catL-hatching gland), opl (telencephalon), en2 (midbrain/hindbrain boundary), krox20 (rhombomeres 3 and 5) and myod (adaxial cells and somites).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development