- Title
-
Probing small ribosomal subunit RNA helix 45 acetylation across eukaryotic evolution
- Authors
- Bortolin-Cavaillé, M.L., Aurélie, Q., Supuni, T.G., Thomas, J.M., Sas-Chen, A., Sharma, S., Plisson-Chastang, C., Vandel, L., Blader, P., Lafontaine, D.L.J., Schwartz, S., Meier, J.L., Cavaillé, J.
- Source
- Full text @ Nucleic Acids Res.
|
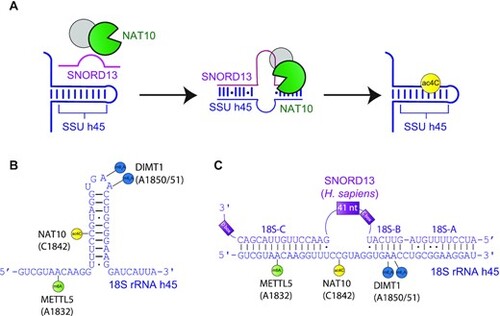
Schematic of SNORD13-dependent ribosomal RNA acetylation. (A) Through an incompletely defined antisense-mediated mechanism, human SNORD13 assists the RNA cytidine acetyltransferase NAT10 in modifying C1842 in the universally conserved helix 45 at the 3? extremity of 18S rRNA. (B) Helix 45 is enriched with post-transcriptional RNA modifications. RNA modifying enzymes and their modified nucleotides (lollipops) are indicated: SSU-ac4C1842 (yellow), SSU-m6A1832 (green) and SSU-m26A1850 and SSU-m26A1851 (blue) are introduced by NAT10, METTL5 and DIMT1, respectively. (C) Human SNORD13 base-pairs with 18S rRNA sequences on each side of the cytidine to be acetylated. Note that SNORD13-rRNA duplexes tolerate G-U wobble base-pairs, mismatches or even bulged nucleotides and the length of the intervening loops, from either rRNA or SNORD side, also differ from one organism to another (Supplementary Figure S1). |
|
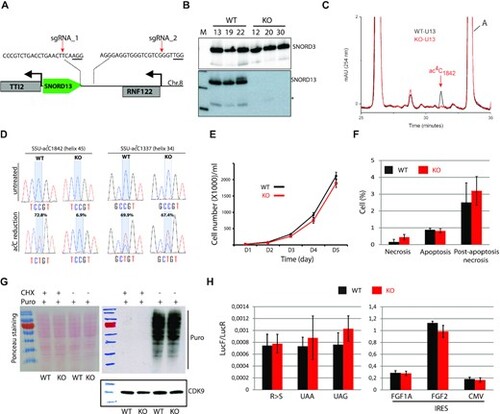
The specific lack of SNORD13-mediated acetylation in helix 45 does not alter cell growth or translation. (A) Schematic representation of SNORD13 locus at human chromosome 8. The relative location of the two DNA sequences targeted by sgRNAs is shown (PAM sequence is underlined). (B) Northern blot showing that SNORD13 is no longer detected in clones bearing deletions. * indicates truncated SNORD13 form which is routinely detected and corresponding to RNA species whose 5?-end is positioned 5?6 nucleotides upstream of the C-box (not shown). (C) RP-HPLC chromatograms of nucleosides obtained from helix 45 of WT (black) and SNORD13-KO (red) cells. Peak corresponding to SSU-ac4C1842 is indicated. (D) Sanger DNA sequencing of RT-PCR products obtained after borohydride treatments of total RNA extracted from WT and SNORD13-KO cells. Percentage of misincorporation (C-to-U) at SSU-C1842 (helix 45) and SSU-C1337 (helix 34) is indicated above each electropherogram. (E) Growth curves of WT and SNORD13-KO cells as judged by cell counting (n = 6 for each genotype). (F) Apoptosis levels determined by flow cytometry using propidium and Annexin V staining of WT and SNORD13-KO cells (n = 2 for each genotype). (G) SUnSET assay. Incorporation of puromycin into the elongating peptide was assayed in WT and SNORD13-KO cells (western blotting with anti-puromycin antibodies). Ponceau S staining of the membrane and detection of CDK9 (western blotting) were used as gel loading controls. Cells treated with the translation inhibitor cycloheximide (CHX) were also used as negative controls. (H) Luciferase reporter gene assays. WT and SNORD13-KO cells were transiently co-transfected by two plasmids expressing Renilla luciferase gene (LucR; internal control) or Firefly luciferase gene (lucF; translation reporter) carrying either an in-frame stop-codons (UAA, UAG), a detrimental mutation (R-to-S) or an IRES from FGF1A, FGF2 and CMV. Histograms show the normalized luciferase activity (lucF-to-lucR ratio). Data are expressed as mean ± s.e.m. and represent 5?6 independent experiments with triplicate measurements. |
|
Phenotypic analyses of SNORD13-deficient zebrafish embryos. (A) Schematic representation of the zebrafish snord13 locus. The relative location of the two DNA sequences targeted by sgRNAs is shown (PAM sequence is underlined). (B) Northern blotting showing that SNORD13 is no longer detected in embryos bearing deletion events. *: truncated SNORD13 form which is routinely detected. SNORD118 was used as a gel loading control. (C) Histograms show percentage of misincorporation (C-to-U ratio) measured in borohydride-treated RNA samples prepared from WT and SNORD13-KO embryos (n = 3). (D?G) Confocal projections of WT (D, F) and SNORD13-KO (E, G) embryos after immunostaining against cleaved Caspase-3 (red) and the pan-neural marker HuC/D (green). (D, E) Dorsal view of the zebrafish brain at 30 hpf, neurons from the olfactory epithelium (oe), tectum (t), pineal gland (pg), optic tectum (ot) and trigeminal ganglion (tg) can be detected. (F, G) Lateral view of the spinal cord (sc) at 30 hpf. The white arrow indicates an apoptotic cell stained by activated caspase-3. (H, I) Confocal projections of WT (H) and SNORD13-KO (I) embryos after in situ RNA hybridization using antisense riboprobes directed against fli1 transcripts. Note that in the absence of antibodies to FLI1 that give reliable signals, fli1 mRNA expression was used as a proxy for monitoring vessel development. It does not imply that fli1 expression per se may be altered in SNORD13-KO zebrafish, whether at the mRNA or protein levels. White brackets indicate the dorsal aorta (DA) and the cardinal vein (CV) and the white arrows point to intersegmental vessels. Pictures are representative of three independent experiments (with at least 6 embryos imaged per genotype). |
|
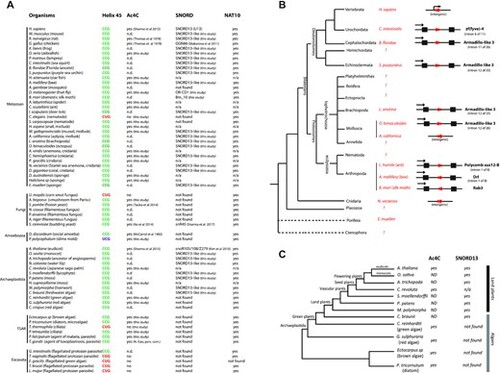
Cross-evolutionary survey of eukaryotic rRNA acetylation machinery. (A) This table summarizes the conservation of the 5?-CCG-3? motif, the acetylation status of helix 45 and the detection of SNORD13 and NAT10 genes in commonly used models or organisms for which we experimentally assayed the presence of ac4C at helix 45. n.d.: not determined. n/a: not applicable (genome is not available) (56,57). (B) Simplified phylogenetic tree of Metazoan. The genomic organization (intronic vs intergenic) of newly-identified SNORD13 genes (red arrows) is shown. Note that intronic SNORD13 can be positioned in the sense or antisense orientation with respect to their host-genes (black arrow). (C) Simplified phylogenetic tree of Archaeplastida. The acetylation status of helix 45 and the presence of SNORD13 in the genome of representative species are indicated. Note that while rRNA in algal species is acetylated, we failed to identify obvious SNORD13 counterparts. A full list of newly-identified SNORD13 sequences can be found in Supplementary data S1. |
|
Atypical SNORD13-related RNA guides rRNA acetylation in D. melanogaster. (A) Simplified phylogenetic tree of Arthropoda. Representative species in which SNORD13-like were identified are indicated. (B) Simplified phylogenetic tree of Diptera. The genomic arrangements of newly-identified SNORD13 genes (blue arrow) are shown. (C) Multiple sequence alignment of representative eukaryotic SNORD13 sequences. The small red arrow indicates the relative position of the mature 5?-end of Or-CD1 in Diptera (see also panel F) while the two horizontal black arrows in opposite orientation depict the terminal 5?-3? stem structure. Note that SNORD13 sequences in Diptera - but not in other species - lack one of the two conserved antisense rRNA elements (denoted by horizontal blue bars). Vertical black, grey and white bars highlight Diptera, other Arthropoda and non-Arthropoda species, respectively. (D) Immunoprecipitation by anti-trimethyl cap (R1131) antibodies. OR-CD1 in various insects (as indicated below the panels) was detected by Northern blots. The same membrane was hybridized with an antisense oligo probe that recognized uncapped 5.8S rRNA used as negative controls. Input RNA (I); RNA recovered from the pellet (P). RNA recovered from the supernatant (S). The theoretical size of SNORD13 (nt) in each of the species studied is indicated in parentheses. (E) Schematic representation of two distinct modes of expression: independent transcription gives rise to short (top) or long (bottom) forms of 5? capped SNORD13. (F) A mutant KO fly strain harboring an inserted P-element in the Or-CD1 gene (Dmel\P{EP}snoRNA:Or-CD1G9117) does not express OR-CD1 as assayed by primer extension. Note that the 5?-end of the cDNA product confirms that fly Or-CD1 lacks one 18S rRNA complementarity. The expected size (nt) of OR-CD1 with ?4?5 nucleotides upstream of the C-box (as shown on the panel) is indicated in parentheses. 32P labeled primer (P). (G) Misincorporation (C-to-U) at SSU-C1968 (helix 45) as judged by Sanger DNA sequencing of RT-PCR products obtained after borohydride treatments of total RNA extracted from WT and Or-CD1-KO adult flies. |
|
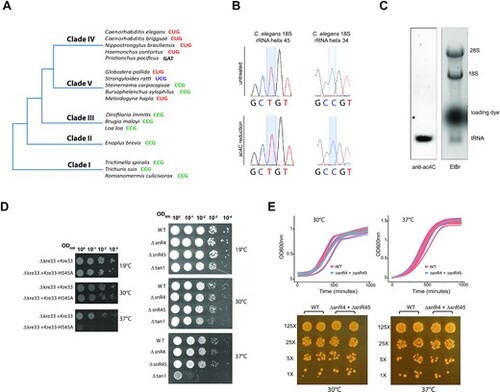
Life without rRNA acetylation (A) Simplified phylogenetic tree of Nematoda. Sequence contexts of the acetylated cytidine in representative species of each clade are indicated. (B) Sanger DNA sequencing of RT-PCR products obtained after borohydride treatments of total RNA extracted from C. elegans. (C) Ac4C content in total RNA extracted from adult C. elegans was visualized by Northern blotting-based assay using anti-Ac4C antibodies. Ethidium bromide (etBr) staining indicates the relative position of rRNA and tRNA species. (D) A ten-fold serial dilution of cell suspensions of the different yeast strains (as indicated on the side of the panels) was spotted on a YPD-agar plate and incubated for 48 h at low (19°C), regular (30°C) and high (37°C) temperatures. (E) Top-panels: WT and ?snR4?snR45 yeast strains were grown at 30°C (left) or 37°C (right) in liquid YPD. Optical density at 600 nm (OD600) was measured every 30 min. Growth curves of individual biological samples (thin lines; n = 3 or 12 for 30°C and 37°C samples, respectively) and the mean of replicates (thick lines) are presented along with a confidence interval (gray area). Bottom-panels: WT and ?snR4?snR45 yeast strains were spotted on a YPD-agar plate and incubated for 48 h at 30°C or 37°C. Cells were grown in duplicates with a 5-fold dilution between distinct concentrations. |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. PHENOTYPE:
|