- Title
-
Lysyl hydroxylase 3 is required for normal lens capsule formation and maintenance of lens epithelium integrity and fate
- Authors
- Taler, K., Weiss, O., Rotem, S., Rubinstein, A.M., Seritrakul, P., Gross, J.M., Inbal, A.
- Source
- Full text @ Dev. Biol.
|
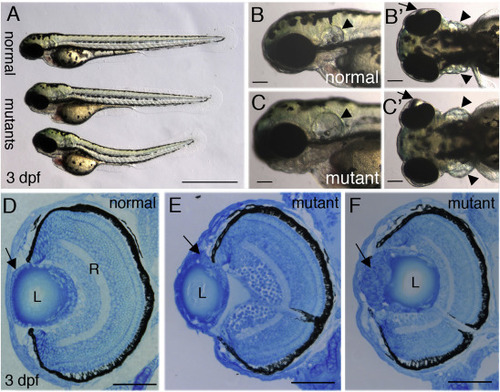
pninavu222 mutant embryos. (A-C’) Live 3 dpf normal and mutant embryos. (A) Lateral view, top embryo is normal, bottom two embryos are mutants (n > 100 embryos). (B,B’,C,C’) Higher magnifications of head region, lateral views (B,C) and dorsal views (B’,C’) of normal (B,B’) and mutant embryos (C,C’). Arrows point at lenses, arrowheads at ears. (D–F) Transverse sections of eyes from normal (D) and mutant (E,F) embryos at 3 dpf (n > 10 for each genotype). Arrow in D points at the ALE and in E,F at abnormal masses of cells. L, lens; R, retina. Scale bars are 1 mm (A) and 50 μm in all other panels. PHENOTYPE:
|
|
Early lens abnormalities in pninavu222 mutants. (A–F) Transverse sections through eyes of normal siblings (A,C,E) and pnina mutants (B,D,F) at different time points, as depicted (n = 10 for each genotype and time point). Asterisks mark the CMZ and arrows point at the ALE. Arrowheads in D point at abnormal regions in the ALE. (G,H) Confocal single-plane images of transverse sections of eyes from normal (G) and mutant (H) 54 hpf embryos, labeled for pHH3 (green) and nuclei (DAPI, blue). Arrowheads and arrows point at pHH3-positive cells in the ALE and CMZ, respectively. (I) Statistical analysis of pHH3 labeling (t-test, one-tailed, error bars show s. e.m). (J,K) Merge of confocal and bright-field images of TUNEL in whole 50 hpf normal (J) and mutant (K) eyes (dorsal view), showing the ALE and lens region. Arrowheads point at TUNEL positive cells and arrows at ALE. (L) Statistical analysis of TUNEL experiment (t-test, one-tailed, error bars show s. e.m). (M,N) Merge of confocal fluorescence and bright-field single-plane images of lenses of Tg(foxe3:nls-EGFP) 3 dpf normal (M) and mutant (N) embryos. Insets show EGFP fluorescence channel alone. EGFP expression (green) is evident in the normal ALE (M, arrows) and in the mutant’s cell mass (N, arrows). EGFP expression is also present in nuclei of surface epithelium (arrowheads). Scale bars in all panels are 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
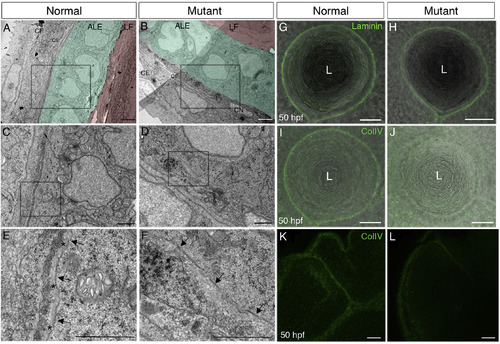
Lens capsule is abnormal in pninavu222 mutants. (A–F) Representative transmission electron microscope (TEM) images of lens epithelium and lens capsule in a normal sibling (A,C,E) and pninavu222/vu222 (B,D,F) embryo at 48 hpf (n = 3 embryos for each genotype). (A,B) The color coding highlights lens fibers (LF) (red), anterior lens epithelium (ALE) (green) and the bi-layered corneal epithelium (CE) (no color). Corneal stroma (CS) and corneal endothelium (CN) are at early stages of formation. C and D are higher magnifications of the regions marked by rectangles in A and B, respectively. E and F are higher magnifications of the regions marked by rectangles in C and D, respectively. Arrows in E, F point at basal cell membranes of anterior LECs and asterisks in E mark the lens capsule. (G–L) Single confocal planes from 50 hpf normal (G,I,K) and mutant (H,J,L) embryos, lateral views, anterior to the left. Labeling (green) is for Laminin (G,H) or ColIV (I–L). Images show the equator region of lenses (G–J) or forebrain and midbrain regions (K,L). Bright-field images in G-J are overlaid to highlight tissue morphology. Numbers of embryos from two repetitions for each staining: G = 7, H = 9, I = 5, J = 7. L, lens. Scale bars: A,B - 2 μm; C–F - 1 μm; G-L - 20 μm. |
|
pnina is allelic to plod3. (A,B) Transverse histological sections through eyes of 3 dpf normal sibling (A) and plod3tv205/tv205 (B) embryos (n = 10 embryos from each genotype). (C,D) Live 3 dpf embryos from a cross between pninavu222/+ and plod3tv205/+ fish. From over 200 embryos, approximately 75% of embryos appeared normal (top embryo) and 25% showed a plod3 mutant-like phenotype (bottom embryo). C is lateral view, D, dorsal view (compare to Fig. 1A,B’,C’). (E) Relative plod3 expression as measured by RT-qPCR in plod3vu222/vu222 mutant and normal embryos. Blue lines show mean expression level (relative quantity, RQ) and bars represent highest and lowest expression levels based on the standard error of the delta Ct’s. (F,G) Transverse sections through eyes of 30 hpf (F) and 48 hpf (G) wild-type embryos labeled for plod3 expression by in situ RNA hybridization (n = 10 eyes for each time point). Arrows in E point at the ALE and in F at CMZs. Scale bars are 50 μm (A,B), 1 mm (C), 100 μm (D), 20 μm (F,G). PHENOTYPE:
|
|
Development of lens cellular mass phenotype. (A) Live imaging of a lens of a ~72 hpf plod3vu222/vu222 embryo carrying a mem-EGFP transgene (n = 5 embryos). Dorsal view, anterior to the top. ALE cells forming a multi-layered cell mass are seen next to the lens. (B–E) Transverse histological sections from eyes of plod3vu222/vu222 embryos at 75 hpf (B), 4 dpf (C,D) and 5 dpf (E) (n = 10 eyes for each time point). Red lines encircle major parts of cellular masses in B-D and a region of the mass forming a rounded pattern in E. L, original lenses. Scale bars are 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cellular masses are disorganized and partially differentiated towards lens fiber fate. (A–H) Single confocal planes of eyes from normal (A,A’,C,E,G) and plod3vu222/vu222 (B,B’,D,F,H) embryos labeled (red) for ZO-1 (A,A’,B,B’), Pax6 (C,D), Prox1 (E,F) and Aq0 (G,H). Nuclei (DAPI) are blue. A’ and B’ are higher magnifications of the regions marked by rectangles in A and B, respectively. Areas of cellular masses are encircled by white lines in D,F,H. Arrows in A point at the ALE and in C at Prox1-positive differentiating lens fiber cells. Insets in C and D show only the Pax6 channel in the region of the ALE and cell mass, respectively. Ages of embryos are demarcated on panels. Number of eyes imaged from two repetitions of the experiments are: A = 9, B = 14, C = 7, D = 11, E = 6, F = 19, G = 15, H = 17. L, lens. Scale bars are 20 μm. EXPRESSION / LABELING:
PHENOTYPE:
|
|
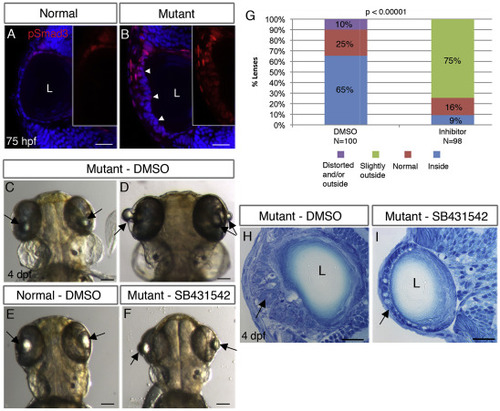
TGFβ signaling drives formation of cellular masses in plod3-mutant lenses. (A,B) Single confocal planes of eyes from 75 hpf normal (A) and plod3vu222/vu222(B) embryos labeled for pSmad3 (red) and nuclei (DAPI, blue). Numbers of eyes analyzed: A = 8, B = 10. Arrowheads in B point at the abnormal ALE that shows expression of pSmad3, and insets in A and B show pSmad3 channel alone in the ALE region. (C–F) Live 4 dpf plod3vu222/vu222 embryos (C,D,F) or normal sibling (E), dorsal views, anterior to the top. Embryos in C,D,E were treated from 30 hpf with vehicle (DMSO) and in F with SB-431542. Arrows point at lenses. (G) Quantification and statistical analysis of treatment of plod3vu222/vu222 with SB-431542 (χ2 test). Location of lenses relative to retina is represented by different colors. (H,I) Transverse histological sections from eyes of plod3vu222/vu222 embryos at 4 dpf. Embryo in H was treated with vehicle and in I with SB-431542. Ten eyes were analyzed in two separate experiments. Arrows in H point at cellular mass and in I at rescued monolayer of ALE with abnormal cells. Scale bars are 20 μm (A,B,H,I) or 100 μm (C–F). EXPRESSION / LABELING:
PHENOTYPE:
|
|
Inhibition of TGFβ signaling restores normal expression of pSmad3, Prox1 and Aquaporin 0. (A–F) Confocal images of sections from eyes of 4 dpf plod3vu222/vu222 embryos treated with vehicle (DMSO) (A,C,E) or with SB-431542 (B,D,F) to inhibit TGFβ signaling via TGF type I receptor. Sections were labeled for pSmad3 (red, A,B), Prox1 (red, C,D) and Aq0 (red, E,F). In all panels, nuclei are labeled with DAPI (blue). Arrows in A,C,E point at abnormal expression of pSmad3, Prox1 and Aq0, respectively, in the cell masses. Data is representative of the following number of eyes: A = 9, B = 11, C = 6, D = 12, E = 7, F = 10. L, lens. Scale bars are 20 μm. |
|
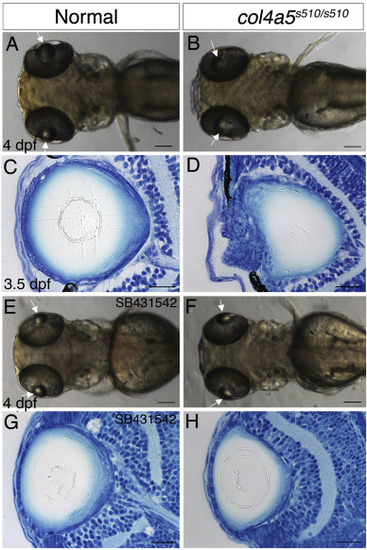
col4a5s510 embryos show the same TGFβ-driven lens phenotype as plod3 mutants. (A,B) Live 4 dpf normal (A) and col4a5s510/s510 (B) embryos. (n > 100 embryos). (C,D) Transverse histological sections of eyes from 3.5 dpf normal (C) and col4a5s510/s510 (D) embryos. (n = 10 eyes from each genotype). (E,F) Live 4 dpf normal (E) (n = 6) and col4a5s510/s510 (F) (n = 5) embryos, treated with SB-431542 from 30 hpf. (G,H) Transverse histological sections from eyes of the same normal and mutant embryos in G and H, respectively. Numbers of eyes analyzed: G = 12, H = 9. A,B,E,F are dorsal views, anterior to left, arrows point at lenses. Scale bars in A,B,E,F are 100 μm and in C,D,G,H 20 μm. PHENOTYPE:
|
Reprinted from Developmental Biology, 458(2), Taler, K., Weiss, O., Rotem, S., Rubinstein, A.M., Seritrakul, P., Gross, J.M., Inbal, A., Lysyl hydroxylase 3 is required for normal lens capsule formation and maintenance of lens epithelium integrity and fate, 177-188, Copyright (2019) with permission from Elsevier. Full text @ Dev. Biol.