- Title
-
microRNA-138 modulates cardiac patterning during embryonic development
- Authors
- Morton, S.U., Scherz, P.J., Cordes, K.R., Ivey, K.N., Stainier, D.Y., and Srivastava, D.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
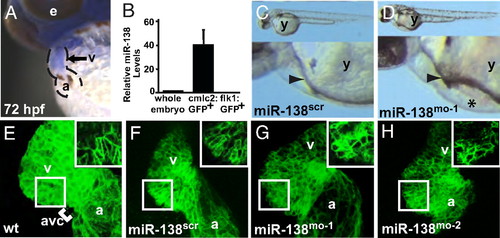
miR-138 is required for cardiac development. (A) Oblique view of 72 hpf fish after in situ hybridization showing expression of miR-138 in the ventricular chamber marked by blue staining. (B) qRT-PCR analysis of mature miR-138 expression at 48 hpf in flow-sorted cardiomyocytes (cmlc2:GFP), endothelial cells (flk1:GFP), or whole embryos demonstrated enrichment of miR-138 in cardiomyocytes. Mature miR-138 was not detected in endothelial cells. (C and D) Fish embryos injected with miR-138 scrambled control morpholino (scr) (C) or miR-138 morpholino (mo-1) (D) showing pericardial edema (asterisk); arrowhead indicates heart. (E–I) Representative confocal images of transgenic Tg(myl7:HRAS-mEGFP)s843 uninjected embryos (E) or embryos injected with 17-nt miR-138scr (F), 17-nt miR-138mo-1 (G), or a second 31-nt miR-138mo-2 oligo (H). Insets show single z-stack images of outer curvature ventricular myocytes, which normally elongate with maturation (E and F) but remained rounded in the miR-138mo embryos (G and H). (e, eye; y, yolk sac; a, atrium; v, ventricle; avc, atrioventricular canal.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
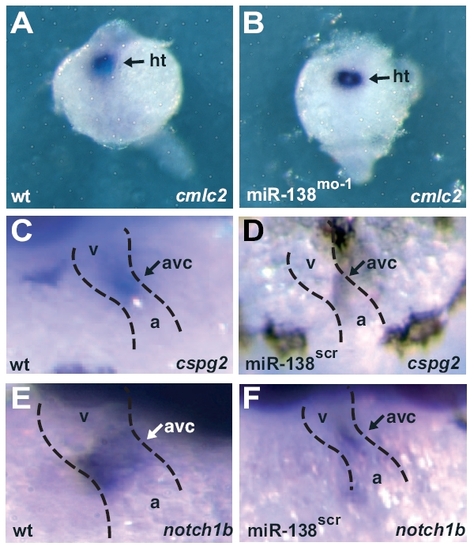
miR-138 knockdown leads to expansion of AVC-specific gene expression into the ventricle. (A–H) Ventral views of 48-hpf embryos after mRNA in situ hybridization focusing on head (h) and heart (dotted lines) regions. Atrial (A and B) and ventricular (C and D) markers were similar in wild-type (wt) (A and C) and miR-138mo-1 embryos (B and D). Expression of AVC-specific markers cspg2 (E and F) and notch1b (G and H) expanded into the ventricles (v) in miR-138mo-1 embryos (F and H) but not in wild-type embryos (E and G). (a, atrium; amhc, atrial myosin heavy chain; vmhc, ventricular myosin heavy chain.) |
|
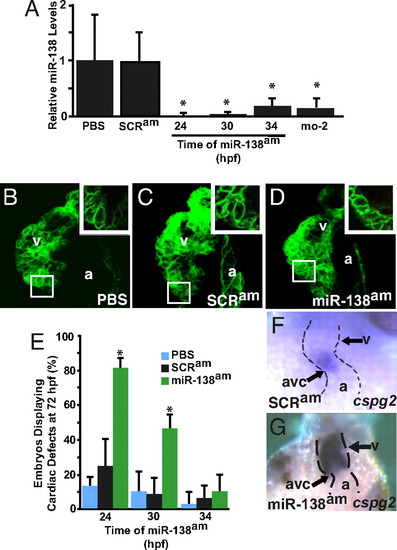
Temporal regulation of miRNA function by antagomiRs in zebrafish. (A) miR-138 RNA levels detected by qRT-PCR in 72-hpf fish embryos treated with antagomiRs from 24, 30, or 34 h postfertilization (hpf) or injected with the 31-nt morpholino (mo-2) compared with PBS- or scrambled antagomiR-treated controls. (B–D) Confocal images of hearts of 72-hpf Tg(myl7:HRAS-mEGFP)s843 transgenic embryos treated with PBS (B), scrambled antagomiR (SCRam) (C), or miR-138 antagomiR (miR-138am) (D) from 24–72 hpf. GFP revealed rounded myocyte morphology (Inset) in miR-138am embryos compared to the normally elongated myocytes seen in controls. (E) Pericardial edema indicating cardiac dysfunction and rounded ventricular myocytes reflecting delayed maturation were observed in varying percentages of 72-hpf embryos treated with PBS, SCRam, or miR-138am from the hpf indicated. (F and G) Ventral view of mRNA in situ hybridization to detect cspg2 expression in hearts of embryos treated with SCRam (F) or miR-138am (G) showed expansion of cspg2 into the ventricle of miR-138am embryos; heart is indicated with dotted lines. Results shown are the average of four experiments in (A) and (E). (v, ventricle; a, atrium; atrioventricular canal (avc); *, P < 0.05.) EXPRESSION / LABELING:
PHENOTYPE:
|
|
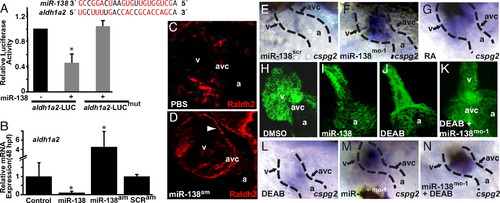
miR-138 directly targets aldh1a2, restricting its expression to the AVC. (A) miR-138 binding site in zebrafish aldh1a2 3′ UTR with complementary nucleotides indicated in red. Luciferase activity in Cos cells on introduction of wild-type or mutated (mut) aldh1a2 3′ UTR sequences downstream of a CMV-driven luciferase reporter with or without miR-138 is shown. (B) aldh1a2 mRNA levels measured by qRT-PCR in embryos injected with pri-miR-138 or treated at 24 hpf with miR-138 antagomiR (miR-138am) or scrambled antagomiR (SCRam). Results shown represent at least 4 experiments in (A) and (B). Error bars indicate 95% confidence intervals. (C and D) Raldh2 immunohistochemistry on sections of 72-hpf embryos treated with PBS or miR-138am from 24–72 hpf. Ventricular (v) cardiomyocyte expression (arrowhead) was readily detectable in miR-138am hearts but not in controls, whereas intensity of staining in the AVC was similar. (E–G) Ventral view of cspg2 expression in the heart by mRNA in situ hybridization in embryos treated with scrambled morpholino (miR-138scr) (E), miR-138mo-1 (F), or retinoic acid (RA) (G); heart is indicated with dotted lines. (H–K) Confocal images of myl7-GFP embryos (48 hpf) treated with vehicle (DMSO) (H), premiR-138 RNA (I), or DEAB (J), and DEAB-treated embryos also injected with miR-138mo-1 (K) showing rescue of the linear heart tube phenotype with knockdown of miR-138. (L–N) Expansion of cspg2 mRNA expression from the avc (L) into the ventricle (v) of miR-138mo-1 embryos (M) was not rescued by DEAB (N). (a, atrium; *, P < 0.05.) |
|
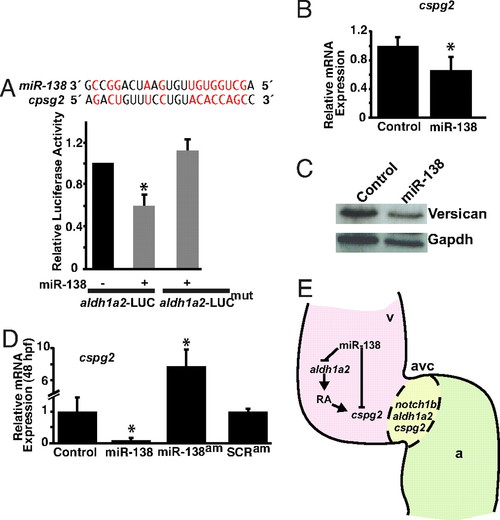
miR-138 directly targets cspg2. (A) Sequence of miR-138 and its binding site in mouse cspg2 3′ UTR with complementary nucleotides indicated in red. Luciferase activity in Cos cells on introduction of wild type or mutated (mut) cspg2 3′ UTR sequences downstream of a CMV-driven luciferase (LUC) reporter with or without miR-138 is shown. (B and C) Analysis of cspg2 mRNA level assessed by qRT-PCR (B) and corresponding versican protein by western blot (C) in mouse NIH 3T3 fibroblasts transfected with miR-138. (D) cspg2 mRNA levels detected by qRT-PCR in zebrafish embryos injected with miR-138 or treated at 24 hpf with miR-138 antagomiR (am) or scrambled (SCR) antagomiR. (E) Proposed model for the function of miR-138 in regulating chamber-specific gene expression and atrioventricular canal (avc) patterning. miR-138 directly represses retinoic acid (RA) synthesis in the ventricle (v) via aldh1a2, which would otherwise induce expression of the avc-specific gene, cspg2 (versican). Repression of cspg2 by miR-138 in the ventricle is also accomplished by directly targeting cspg2. Results shown in (A), (B), and (D) represent at least 4 experiments with error bars indicating 95% confidence intervals. (a, atrium; *, P < 0.05.) |
|
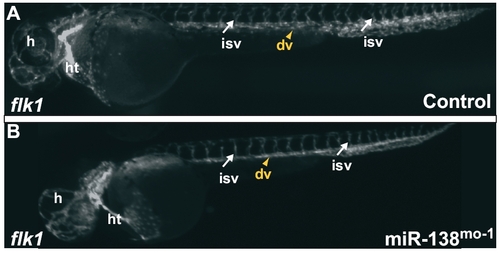
(A and B) Tg(flk1:EGFP)s843 embryos, where GFP expression at 48 hpf marks endocardial cells and endothelial cells in blood vessels. Blood vessels between the somites and within the head appear similar in control (A) and miR-138-morpholino injected fish (B). (dv, dorsal vessels; h, head; ht, heart; isv, intersomitic vessels.) |
|
(A and B) Expression of cmlc2 in dorsal views at 24 hpf as determined by in situ hybridization (purple) in whole-mount embryos indicates that cardiac progenitors are similarly specified in WT control (A) and 138-morpholino-injected fish (B). (C–F) In situ hybridization for cspg2 (C and D) and notch1b (E and F) in wild-type (C and E) and miR-138scr fish (D and F) demonstrate atrioventricular-specific expression at 48 hpf. a, atrium; ht, heart progenitors; v, ventricle. |
|
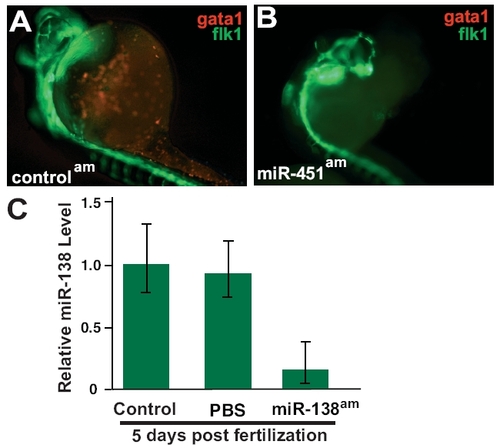
(A and B) Tg(flk1:EGFPs843; gata1:dsRed)s843 embryos at 32 hpf, where RFP expression marks red blood cells (RBCs) and GFP expression marks endothelial cells. Fewer RBCs were seen in miR-451am embryos (B) than in control antagomiR-treated embryos (A). (C) qRT-PCR analysis of 5-day postfertilization embryos treated with PBS or miR-138 antagomiR demonstrates continued knockdown of miR-138. |
|
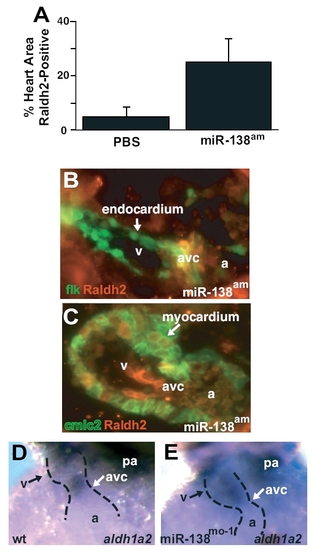
(A) Quantification of Raldh2-positive area in multiple immunohistochemistry sections as represented in main text Fig. 4 C and D. Results shown are the average of 4 slides per group. (B and C) Immunohistochemistry for Raldh2 protein in miR-138am fish demonstrates Raldh2 expression (red) overlapping with Tg(flk1-EGFP)s843 (B) in the atrioventricular canal (avc), and Raldh2 overlapping with Tg(cmlc2-ras-EGFP)s843 in the ventricle (v) (C). cmlc2-ras-EGFP expression is localized to the cell membrane of cardiomyocytes. (D and E) In situ hybridization for aldh1a2 demonstrates AVC-restricted expression in control fish (D) but expanded ventricular expression in miR-138mo fish (E). wt, wild-type; pa, pharyngeal arch; a, atrium. |