Fig. 1
Fig. 1
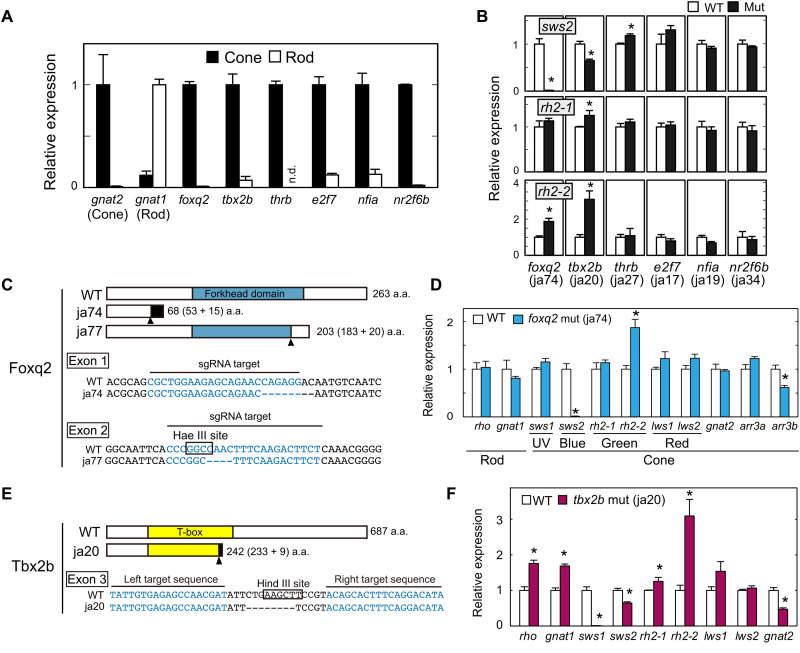
(A) Relative expression levels of phototransduction genes and transcription factors in isolated rods and cones at the adult stage (means ± SD, n = 2). n.d., not detected. The expression levels of gnat1 and gnat2 genes are reproduced from our previous paper (17). (B) Relative expression levels of sws2 opsin in the larval eyes at 5 days postfertilization (dpf). Means ± SEM. *P < 0.05 by Student’s t test. The number of fish used was as follows: n = 5 [foxq2 wild type (WT)], n = 5 (foxq2 mut); n = 5 (tbx2b WT), n = 5 (tbx2b mut); n = 3 (thrb WT), n = 4 (thrb mut); n = 4 (e2f7 WT), n = 4 (e2f7 mut); n = 3 (nfia WT), n = 4 (nfia mut); n = 4 (nr2f6b WT), n = 4 (nr2f6b mut). See also fig. S3. (C and E) Schematic representation of Foxq2 and Tbx2b and their partial nucleotide sequences. The frameshift site is indicated by an arrowhead. Nucleotide deletions are indicated by dashes. The nucleotide sequences (letters in blue) indicate the target sequences of TAL effector nucleases or Cas9–single-guide RNA (sgRNA) complexes. The recognition sites of the restriction endonucleases, Hae III and Hind III, are surrounded by black lines. The ja74, ja77, and ja20 mutations caused a frameshift of the amino acid (a.a.) sequence of Foxq2 or Tbx2b by 8-, 4-, and 8-bp loss, respectively. (D and F) Expression profiles of phototransduction genes in the 5-dpf larval eyes. Means ± SEM. *P < 0.05 by Student’s t test. The number of fish used was as follows: n = 5 (foxq2 WT), n = 5 (foxq2 mut); n = 5 (tbx2b WT), n = 5 (tbx2b mut). The expression levels of sws2 and rh2 genes are reproduced in (B). UV, ultraviolet.