Fig. 5 - Supplemental 1
- ID
- ZDB-FIG-250722-74
- Publication
- Wang et al., 2025 - The geometry and dimensionality of brain-wide activity
- Other Figures
-
- Fig. 1
- Fig. 2
- Fig. 2 - Supplemental 1
- Fig. 2 - Supplemental 2
- Fig. 2 - Supplemental 3
- Fig. 2 - Supplemental 4
- Fig. 3
- Fig. 3 - Supplemental 1
- Fig. 3 - Supplemental 2
- Fig. 4
- Fig. 4 - Supplemental 1
- Fig. 4 - Supplemental 2
- Fig. 5
- Fig. 5 - Supplemental 1
- Fig. 5 - Supplemental 2
- Fig. 5 - Supplemental 3
- Fig. 5 - Supplemental 4
- Fig. 5 - Supplemental 5
- Fig. 5 - Supplemental 6
- Fig. 5 - Supplemental 7
- Fig. Appendix Figure 1
- Fig. Appendix Figure 1 (2)
- Fig. Appendix Figure 2
- Fig. Appendix Figure 2 (2)
- Fig. Appendix Figure 3
- Fig. Appendix Figure 3 (2)
- Fig. Appendix Figure 4
- Fig. Appendix Figure 5
- Fig. Appendix Figure 6
- Fig. Appendix Figure 7
- Fig. Appendix Figure 8
- Fig. Appendix Figure 9
- All Figure Page
- Back to All Figure Page
|
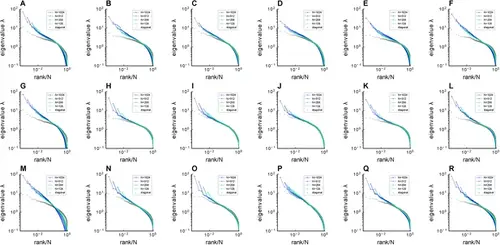
Fitting Euclidean Random Matrix (ERM) to zebrafish data from our experiments (part 1). Comparison of sampled covariance eigenspectra in fish data and fitted ERM models. The columns correspond to six light-field zebrafish data: fish 1 to fish 6. Number of time frames: fish 1 – 7495, fish 2 – 9774, fish 3 – 13,904, fish 4 – 7318, fish 5 – 7200, and fish 6 – 9388. (A–F) sampled covariance eigenspectra for different fish data. (G–L) Same as (A–F) but for ERM models with fitted parameters (μ/d , L ), functional coordinates inferred using MDS, and the experimental σi . (M–R) Same as (A–F) but for ERM models with fitted parameters (μ/d , L ), uniform distributed functional coordinates, and a log-normal distribution of σ2 . μ/d=[0.456,0.258,0.205,0.262,0.302,0.308] in fish 1–6. |