Fig. 5
- ID
- ZDB-FIG-250428-5
- Publication
- Thiruppathy et al., 2025 - Repurposing of a gill gene regulatory program for outer ear evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
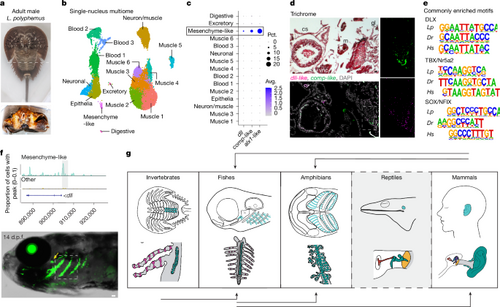
a, Adult male L. polyphemus with a dashed line indicating the dissection plane. Book gills (bottom) showing the region dissected for multiome (dashed box). b, Book-gill clusters annotated by cell type. c, Dot plot showing dll, comp-like and alx1-like expression in the mesenchyme-like cluster. Pct., percentage; Avg., average. d, Trichrome staining of a frontal section of a juvenile one-year-old horseshoe crab shows cardiac sinus (cs), medial midgut (mg), ventral nerve cord (vnc), lateral gill lamellae (gl), cartilage-like tissue (c) and muscle (m). RNA in situ hybridization of an adjacent section shows co-expression of dll and comp-like in mesenchyme-like cells at the growing edge of the cartilage (single channels on the right show the magnified box region). DAPI labels nuclei. e, Of 27 de novo motifs enriched in DARs from horseshoe crab (Lp) mesenchyme-like cells, 3 are conserved in DARs of zebrafish (Dr) gill cartilage and human (Hs) outer-ear cartilage. f, Top, genome coverage plot shows the differential accessibility of Lp_dll-p1 in mesenchyme-like cells. Bottom, Lp_dll-p1:GFP drives transgenic activity in the gills (boxed region) and in the gill-like pseudobranch (yellow arrow) at 14 d.p.f. g, Evolutionary model for the redeployment of the ancestral gill program across species. Top, putative locations of gill program-derived structures (turquoise) in invertebrates (horseshoe crab book gills), fishes (zebrafish gills), amphibians (Xenopus tadpole gills), reptiles (anole ear cavity) and mammals (human outer ear). Bottom, details of the structures, highlighting the elastic cartilage (green), respiratory cells (red) in the secondary lamellae of crabs and fishes, and ear structures of anoles and humans (columella/stapes, red; incus, blue; malleus, purple; tympanic membrane, yellow; outer ear canal, grey). Black arrows show where cross-species enhancer testing supports gill-program redeployment. For anoles, the dashed border and grey background denote a lack of enhancer testing. Scale bar, 50 µm. Credits: g, created by M.T. in BioRender (https://BioRender.com/c72e915, https://BioRender.com/p21u076, https://BioRender.com/q06o675, https://BioRender.com/v80c208, https://BioRender.com/t94e268, https://BioRender.com/b31e815, https://BioRender.com/c97i419 and https://BioRender.com/l10k434). |