Fig. Ext. 10
- ID
- ZDB-FIG-250428-15
- Publication
- Thiruppathy et al., 2025 - Repurposing of a gill gene regulatory program for outer ear evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
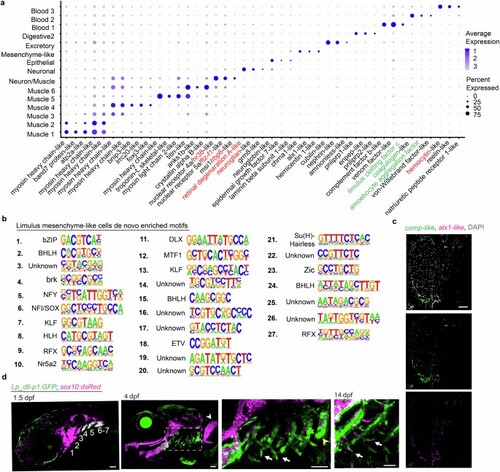
a, Dot plot of genes from top 10 differentially expressed for each cluster. Gene names assigned based on predicted homology of their encoded proteins with vertebrate or invertebrate proteins as determined using blastp. Those in black are from predicted homology with vertebrate proteins, in red from predicted homology with Drosophila proteins, and in green proteins that have been characterized in Limulus species. b, De novo motif enrichment analysis conducted on DARs identified from the cluster of horseshoe crab mesenchyme-like cells. c, RNA in situ hybridization for comp-like and alx1-like genes on a frontal section of a juvenile horseshoe crab book gill cartilage show co-enrichment in mesenchyme-like cells along the growing tip of the cartilage. Merged channel with DAPI to highlight nuclei is shown at top, with individual channels below. d, Lp_dll-p1:GFP drives transgenic reporter activity in the gill-forming posterior pharyngeal arch CNCCs (4–7) at 1.5 dpf, and in the base and developing filaments of gills (boxed region) at 4 and 14 dpf. Transgene activity is also seen in the tooth-forming seventh arch (yellow arrowhead), but not in the pectoral fin (white arrowhead). sox10:dsRed labels CNCCs at 1.5 dpf and cartilage at 4 and 14 dpf. Scale bars, 50 µm. |