Fig. 2
- ID
- ZDB-FIG-250428-2
- Publication
- Thiruppathy et al., 2025 - Repurposing of a gill gene regulatory program for outer ear evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
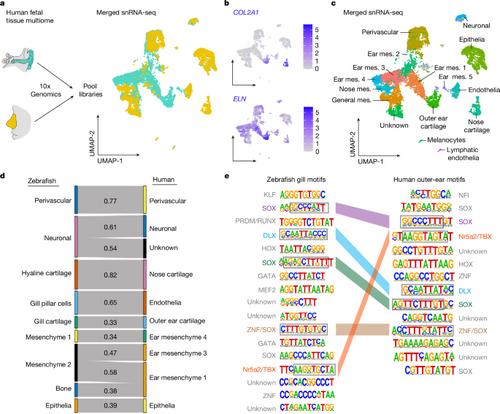
a, Scheme for human 22-week-old fetal tissue dissection, multiome and data analysis, with uniform manifold approximation and projections (UMAPs) of merged outer ear (turquoise) and nose (yellow) cells. b, Feature plots show human COL2A1 and ELN mRNA expression. c, Cluster identities for the merged snRNA-seq datasets; mes., mesenchyme. d, Sankey plot showing SAMap homology mapping of 14 d.p.f. zebrafish CNCC clusters and human fetal outer-ear and nose clusters. An alignment score threshold of 0.3 was used, with 1.0 representing perfect homology. e, All significantly enriched de novo DNA motifs in zebrafish gill versus hyaline cartilage and human outer ear versus nose cartilage DARs, and their putative transcription factor family binding. Coloured bars connect similar motifs between species, with common sequences in grey boxes. Credit: a, created by M.T. in BioRender (https://BioRender.com/e08f213 and https://BioRender.com/a53a784). |