Fig. 4
- ID
- ZDB-FIG-250428-4
- Publication
- Thiruppathy et al., 2025 - Repurposing of a gill gene regulatory program for outer ear evolution
- Other Figures
- All Figure Page
- Back to All Figure Page
|
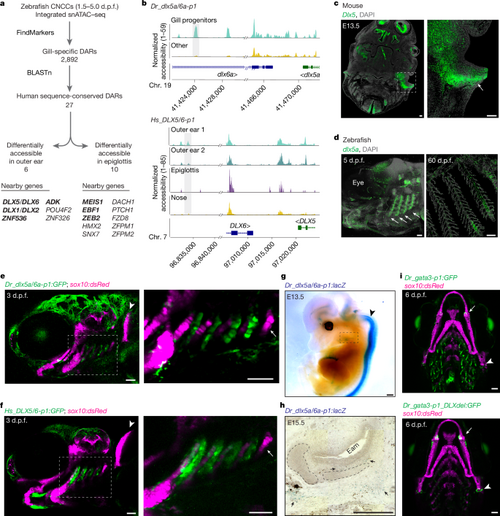
a, Workflow to identify gill developmental DARs with sequence conservation in humans. Genes with confirmed outer-ear or epiglottis expression of mouse homologues are shown in bold. b, Genome coverage plots show differential accessibility of Dr_dlx5a/6a-p1 in 3 d.p.f. zebrafish gills (top) and sequence-conserved region Hs_DLX5/6-p1 in human fetal outer ear (bottom). c, RNA in situ hybridization for Dlx5 in horizontal sections from an E13.5 mouse head. The magnified box region highlights expression in the outer-ear mesenchyme (arrow). d, RNA in situ hybridization for dlx5a in whole-mount zebrafish larva at 5 d.p.f. and in a section through zebrafish gills at 60 d.p.f. Arrows indicate expression in developing gills. Nuclei are labeled by DAPI. e,f, Dr_dlx5a/6a-p1:GFP (e) and Hs_DLX5/6-p1:GFP (f) drive similar reporter activity in the developing gills (boxed region, enlarged on the right) of zebrafish at 3 d.p.f. No activity of either transgene is seen in the tooth-forming seventh arch (arrow) or the pectoral fin (arrowhead). g, Whole-mount X-gal staining of an E13.5 Dr_dlx5a/6a-p1:lacZ mouse. Strong expression is seen in the neural tube (arrow) with weaker expression in the developing ear (boxed region). h, Sagittal section through a whole-mount X-gal-stained E15.5 Dr_dlx5a/6a-p1:lacZ mouse. Arrows indicate positive expression in the mesenchyme surrounding the ear-canal cartilage. Eam, external auditory meatus. i, Wild-type Dr_gata3-p1:GFP strongly labels the gills; Dr_gata3-p1_DLXdel:GFP shows greatly reduced gill activity but retains expression in the jaw joints (arrows) and hyomandibular–opercular joints (arrowheads); sox10:dsRed labels the cartilage. Scale bars: 100 µm in c, g and h; 50 µm in d–f and i. |