- Title
-
Unexpected Phenotype Reversion and Survival in a Zebrafish Model of Multiple Sulfatase Deficiency
- Authors
- Fleming, A., Xuan, L.Z., Sanchez-Elexpuru, G., Williams, S.V., Windell, D., Gelb, M.H., Herbst, Z.M., Schlotawa, L., Rubinsztein, D.C.
- Source
- Full text @ Front Cell Dev Biol
|
FIGURE 1. Protein alignment and mutation sites in human and zebrafish SUMF1/FGE. Human and zebrafish protein sequences share 59.94% sequence identify, increasing to 71.9% over the PFAM domain for FGE-sulfatase activity (yellow highlight). The viral insertion in the sumf1_la015919Tg line occurs in exon one and results in a premature stop codon (blue label). The point mutation in the sumf1_sa31531 line occurs in exon 2 and results in a nonsense mutation and premature stop codon (purple label). |
|
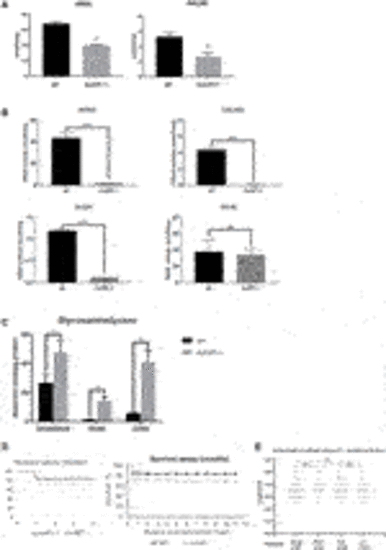
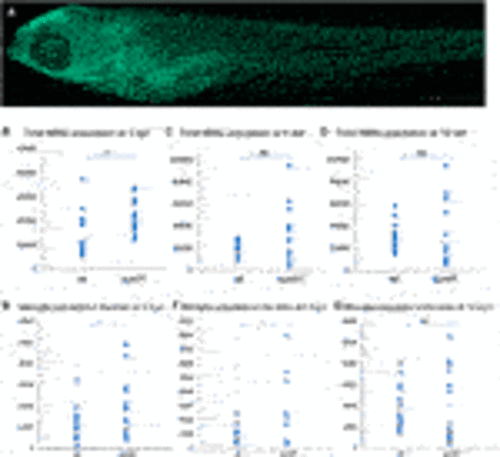
FIGURE 2. Sulfatase activity, GAG accumulation and viability of sumf1−/− zebrafish. (A) Activities of arylsulfatase A (ARSA) and N-acetylgalactosamine-6-sulfatase (GALNS), enzymes that are dependent on FGE function for their activation. Enzymatic activities were measured at 5 d.p.f. in larvae from in-crosses of sumf1_LA+/- adults. A significant reduction in sulfatase activity was confirmed in sumf1−/− larvae, indicating a loss of FGE function. (B) Since larvae from heterozygous females may retain maternal mRNA and/or protein, enzymatic activity assays were performed on sumf1−/− larvae from sumf1−/− parents and compared to wildtype larvae from a wildtype (TL) background. No or negligible levels of ARSA, GALNS, and N-Sulfoglucosamine sulfohydrolase (SGSH) activity could be detected in sumf1−/− larvae at 5 d.p.f. indicating an absence of FGE activity. Beta-galactosidase was included as a control as this enzyme is a lysosomal hydrolase which is not dependent on FGE. (C) Glycosaminoglycan levels were significantly elevated in sumf1−/− larvae at 5 d.p.f. relative to wildtype larvae of the same age. Chondroitin/Dermatan Sulfate (CS/DS) and Heparan Sulfate (HS) apparent levels were calculated using Chondrosine as an internal standard. D0a4: Chondroitin/Dermatan Sulfate Internal Disaccharide 4-sulfation (CS-A, DS) = unsaturated UA-GalNAc (4S); D0a6: Chondroitin Sulfate Internal Disaccharide 6-sulfation (CS-C) = unsaturated UA-GalNAc (6S); D0A0: Heparan Sulfate Internal Disaccharide n-sulfated = unsaturated UA-GlcNS; D0S0: Heparan Sulfate Internal Disaccharide no sulfation = unsaturated UA-GlcNAc. Apparent pmols were calculated using Chondrosine as an internal standard for mass spectrometry-based detection. (D,E) sumf1−/− zebrafish and wildtype fish were reared concomitantly and initially assessed weekly (for 1 month) and then monthly to assess survival. Although an initial drop in viability was observed in sumf1−/− larvae at 14 d.p.f., no further differences in viability were observed up to 12 months of age (D) and no differences in body length were observed in either sex at 30 d.p.f., suggesting that growth was normal (E). For (A–C), graphs show mean values (±SD) from at least 3 biological replicates. Statistical analysis was performed using two-tailed t-test. p > 0.05, *: p ≤ 0.05, **: p ≤ 0.01, and ***: p ≤ 0.001. |
|
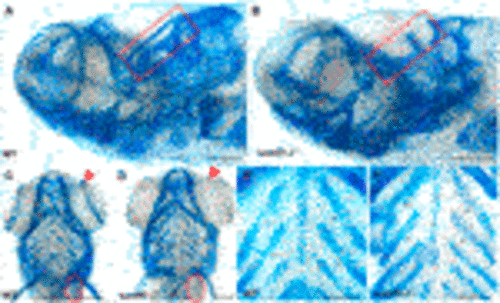
FIGURE 3. Intercalation of chondrocytes in the craniofacial skeleton. Alcian blue stained samples of (A,B) wildtype (n = 4) and (D,E) sumf1−/− zebrafish (n = 6) at 5 d.p.f. The chondrocytes of wildtype larvae (B) at 5 d.p.f. were neatly stacked (asterisk) indicating complete intercalation whereas intercalation in sumf1−/− larvae (E) was disrupted in some areas (outlined) and stacked elsewhere (red star). (C,F). At 10 d.p.f. (C&F, n = 3 per genotype), the intercalation of chondrocytes was disrupted in (F) sumf1−/− larvae (asterisk and red box) when compared to (C) wildtype (asterisk). Scalebar represents 200 μm. |
|
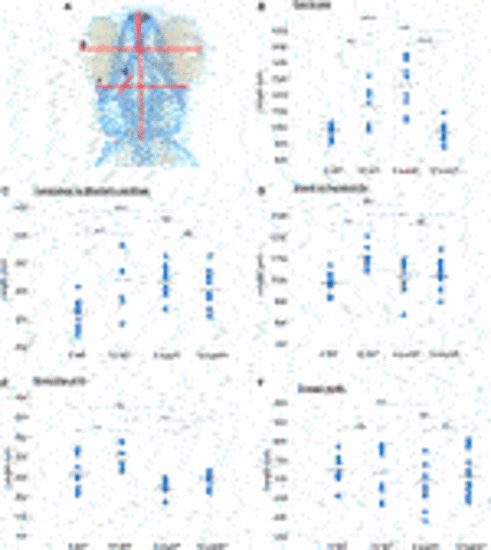
FIGURE 4. Cranial cartilage measurements of wildtype and sumf1 mutant zebrafish at 5 and 10 d.p.f. (A) Schematic diagram indicating measurements of cartilage element representing the following: The distance between the eyes (B), Meckel’s cartilage and ceratohyal (C), snout and the pectoral fin (D), the length of the first branchial arch (E), cranial width (F). Differences in the mean of each measurement was analysed using Welch’s t-test (wildtype and sumf1−/−; 5 d.p.f. and 10 d.p.f.) and the p-values are denoted as following: ns: p > 0.05, *: p ≤ 0.05, **: p ≤ 0.01, and ***: p ≤ 0.001. Means values are indicated by the line. |
|
FIGURE 5. Analysis of craniofacial cartilage at 30 d.p.f. The semi-circular canal (red box) and the scapulocorocoid (red circle) were present in wildtype (A,C) were missing in sumf1−/− larvae (B,D). Similarly, supraorbital cartilage (red arrow) was well developed in wildtype larvae (C) but retarded in sumf1−/− larvae. Notably, chondrocytes in the branchial arches were neatly stacked (intercalated) in wildtype larvae (E), whereas the intercalation of the chondrocytes was disrupted in sumf1−/− larvae (F). Scalebar represents 200 μm. Representative images of n = 3 samples for each genotype. PHENOTYPE:
|
|
FIGURE 6. Development of ossification centres in different craniofacial elements. (A,B) Alizarin red stained larvae were imaged at 10 and 15 d.p.f. and scored according to the size of the ossification centre (growth) and intensity of the fluorescent stain. Ossification centres in the vertebrae (vt), otoliths (oto), primary ossification center (oc) for the hyomandibular (hyo), and ceratohyal (ct), maxilla (m), dentary (d), cleithrum (cl), posterior branchiostegal ray (pbr), ceratobranchial 5 (cb5), operculum (op), entopterygoid (en), branchiostegal ray (bcr) 1 and 2 were assessed (see Table 2). At 30 d.p.f., no overt differences in the mineralisation/ossification of the craniofacial skeleton could be identified between wildtype (C,E) and sumf1−/− larvae (D&F). No differences were found in the fluorescent intensity of the operculum (white box) and ceratohyal (white arrow). The scalebar represents 500 μm. Figure shows representative images of n = 3 samples for each genotype. PHENOTYPE:
|
|
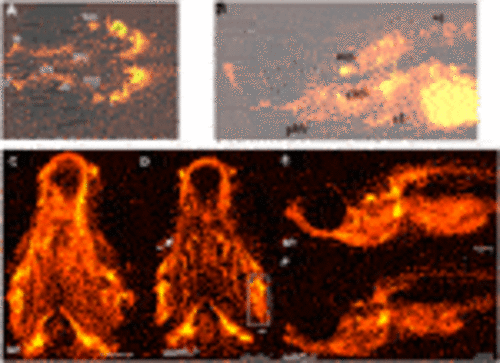
FIGURE 7. Quantification and distribution of microglia and macrophages. (A) Wildtype and sumf1−/− larvae were immunostained with an antibody to detect microglia and macrophages. Scalebar represents 200 μm. The total number of microglia and macrophages (Mi/Ma) was quantified at 3, 5, and 10 d.p.f. by analysing maximum intensity projections of a Z-stack through the entire larva. (B) At 3 d.p.f., there was a significant increase in the Mi/Ma population in sumf1−/− larvae compared to wildtype larvae. However, these differences did not persist at (C) 5 d.p.f. and (D) 10 d.p.f. (E–G) The brain resident population of labelled cells (assumed to by microglia) was quantified at 3, 5, and 10 d.p.f. The brain was selectively quantified by selecting the same Region of Interest (ROI) targeting the frontal and the dorsal brain in all images. There was a significant increase in the microglia population in sumf1−/− larvae at (E) 3 d.p.f. and (F) 5 d.p.f. but no difference at 10 d.p.f. (G). Two-tailed t-test with p-values are denoted as follows: ns: p > 0.05, *: p ≤ 0.05, **: p ≤ 0.01, and ***: p ≤ 0.001. Means values are indicated by the line. PHENOTYPE:
|
|
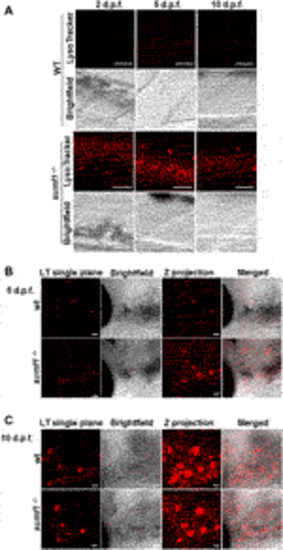
FIGURE 8. Imaging of lysosomes. In vivo lysotracker staining was imaged using confocal microscopy. (A) A striking increase in lysotracker staining was observed in the spinal cord of sumf1−/− larvae compared to wildtype (wt) larvae at 2 d.p.f., 5 d.p.f., and 10 d.p.f. (B) An increase in lysotracker staining was observed in and around the otic vesicle of sumf1−/− larvae compared to wt larvae at 5 d.p.f., whereas no differences between the two groups were observed at 10 d.p.f. (A–C) Representative images of the lysotracker staining, brightfield and the z projection. At least 5 fish were analysed for each group. Scale bar represents 30 µm for all images. |
|
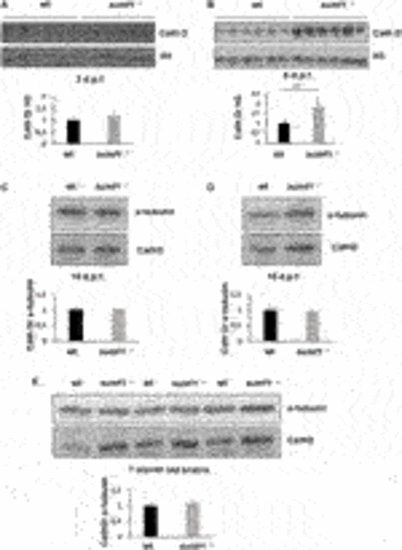
FIGURE 9. Cathepsin D protein levels as an indicator of lysosome digestive capacity. Western blotting was performed in larvae of different ages (3, 5, 10, and 15 d.p.f.) and on brains of adult zebrafish to measure the levels of the mature form of cathepsin D (37 kDa band presented) as an indicator of the digestive capacity of lysosomes. (A) No differences in cathepsin D were observed between wildtype (wt) and sumf1−/− fish at 3 d.p.f. The levels of cathepsin D are significantly increased at 5 d.p.f. in the sumf1−/− fish relative to wt fish (B) whereas no differences were observed between the different genotypes at 10 d.p.f. (C), 15 d.p.f. (D) and in the brains of adult fish (7 months old), (D). Graphs show mean values (± SEM) of densitometry of cathepsin D normalised to histone 3 (loading control at 3 d.p.f and 5 d.p.f.) and α-tubulin (loading control at 10 d.p.f. and 15 d.p.f. larvae and 7 months old brain) from at least 3 independent experiments. Statistical analysis was performed using a two-tailed t-test. ***p < 0.001. PHENOTYPE:
|
|
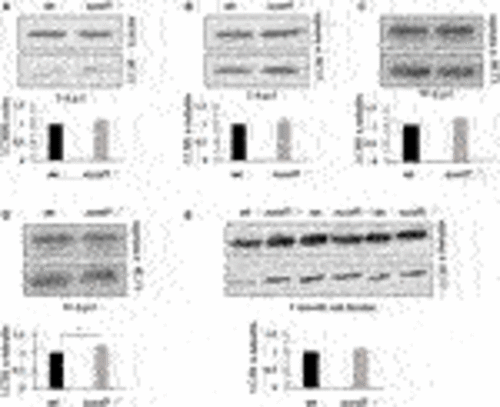
FIGURE 10. LC3-II protein levels as an indicator of autophagosome formation and flux. Western blotting was performed in larvae of different ages (3, 5, 10, and 15 d.p.f.) and on brains of adult zebrafish to measure the levels of LC3-II as a measure of autophagosome number. No differences in LC3-II levels were observed between wildtype (wt) and sumf1−/− fish at 3 d.p.f., 5 d. p.f., 10 d.p.f. Although a modest increase (1.28x) increase in LC3-II was observed at 15 d.p.f. this difference was not evident in the brains of adult fish (7 months). Graphs show mean values (±SEM) of densitometry of LC3-II normalised to β-actin (loading control at 3 d.p.f) and α-tubulin (loading control at 5 d.p.f., 10 d.p.f., 15 d.p.f. and 7 months) from at least 3 independent experiments. Statistical analysis was performed using a two-tailed t-test. *p < 0.05. PHENOTYPE:
|
|
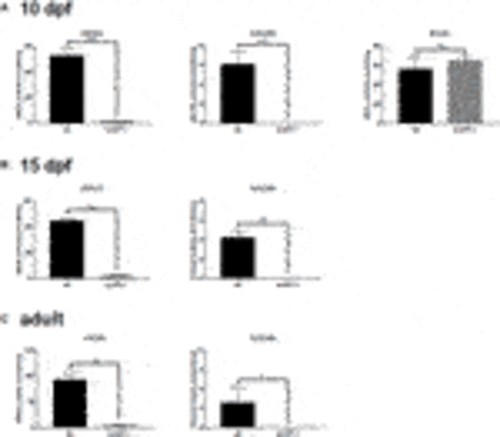
FIGURE 11. Sulfatase activity in older larvae and adult brains. Larvae at 5 d.p.f. showed no or neglible sulfatase activity (Figure 2B). However, since pathological phenotypes in sumf1−/− larvae resolved with age and adult sumf1−/−fish were viable, we measured sulfatase activity at older timepoints. No or negligible levels of ARSA and GALNS activity could be detected in sumf1−/− larvae at 10 and 15 d.p.f., and in the brains of adult fish. Beta-galactosidase was included as a control as this enzyme is a lysosomal hydrolase which is not dependent on FGE. Graphs show mean values (±SD) from at least 3 biological replicates. Statistical analysis was performed using a two-tailed t-test. p > 0.05, *: p ≤ 0.05, **: p ≤ 0.01, and ***: p ≤ 0.001. |