- Title
-
Prmt5 promotes vascular morphogenesis independently of its methyltransferase activity
- Authors
- Quillien, A., Gilbert, G., Boulet, M., Ethuin, S., Waltzer, L., Vandel, L.
- Source
- Full text @ PLoS Genet.
|
(A) Schematic representation of the sequence targeted by CRISPR/Cas9 leading to a 28 nucleotides deletion, and of wild type and truncated Prmt5 proteins. The catalytic domain “CAT” appears in magenta. (B, C) Confocal sections of immunostaining with anti-Prmt5 antibody of wild type and |
|
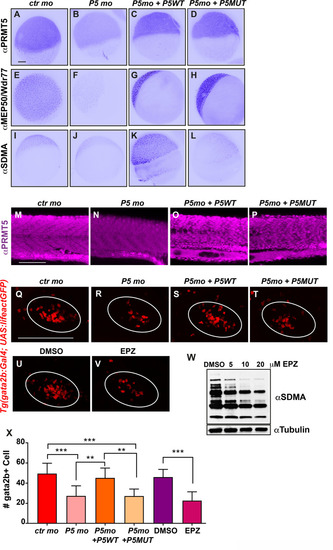
(A-D) PRMT5 immunostaining of 5 hpf embryos injected by control morpholino (A), or prmt5 morpholino only (B) or in combination with prmt5WT mRNA (C) or with the catalytic mutant form prmt5MUT mRNA (D). (E-H) Immunostaining of embryos as in A-D with an antibody raised against MEP50/Wdr77 PRMT5 obligate co-factor. (I-L) Immunostaining as in A-D but with an antibody recognizing SDMAs (symmetric Di-Methyl Arginine motifs) which are PRMT5 substrates. Scale bar, 100 μm. (M-P) Confocal projections of Prmt5 immunostaining of embryos injected by control morpholino (M), or prmt5 morpholino only (N) or in combination with prmt5WT mRNA (O) or with the catalytic mutant form prmt5MUT mRNA (P) at 24 hpf. Scale bar 100 μm. (Q-T) Confocal projections of thymus rudiment from transgenic Tg(gata2b:Gal4; UAS:lifeactGFP) embryos at 3 days in the same conditions as in M-P. Thymus is delimited by a white circle. (U-V) Confocal projections of thymus rudiment from transgenic Tg(gata2b:Gal4; UAS:lifeactGFP) embryos treated with either DMSO (U) or 20 μM of PRMT5 inhibitor EPZ015666 (V) from 30 hpf to 3 dpf. Thymus is delimited by a white circle. Scale bar 100 μm. (W) Western blot of protein extracts from 24 hpf embryos treated either with DMSO or with increasing concentrations (5–20 μM) of EPZ015666, revealed with an antibody recognizing SDMAs. Reblotting with α tubulin antibody served as a loading control. (X) Average number of HSPCs enumerated at 3 days per confocal stack in injected embryos as in Q-V. Data are from 2 independent experiments with at least 3 individuals per analysis. T-test was performed. **P<0.01. ***P<0.001. EXPRESSION / LABELING:
PHENOTYPE:
|
|
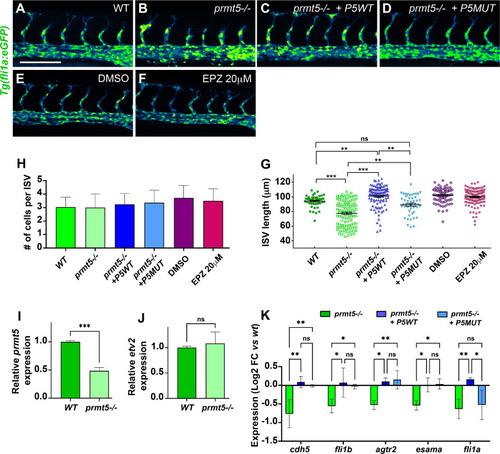
(A-D) Confocal projections of transgenic |
|
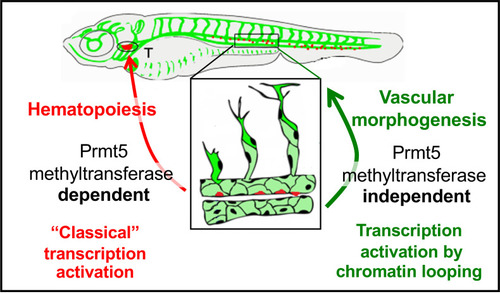
Schematic representation of the two distinct roles of Prmt5 during the formation of hematopoietic lineages and blood vessels, relying or not on its methyltransferase activity, respectively. |
|
(A) Schematic representation of the trunk vasculature with Intersegmental Vessels (ISV) sprouting from the dorsal aorta (DA). The tip cell leads the cell migration and the stalk cell maintains the connection with the DA. CV: cardinal vein. (B, C) Confocal projections of transgenic |
|
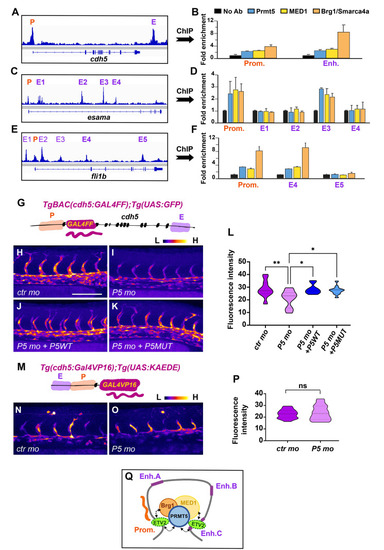
(A-F) ChIP experiments with the indicated antibodies on |