- Title
-
Live Imaging of Heart Injury in Larval Zebrafish Reveals a Multi-Stage Model of Neutrophil and Macrophage Migration
- Authors
- Kaveh, A., Bruton, F.A., Buckley, C., Oremek, M.E.M., Tucker, C.S., Mullins, J.J., Taylor, J.M., Rossi, A.G., Denvir, M.A.
- Source
- Full text @ Front Cell Dev Biol
|
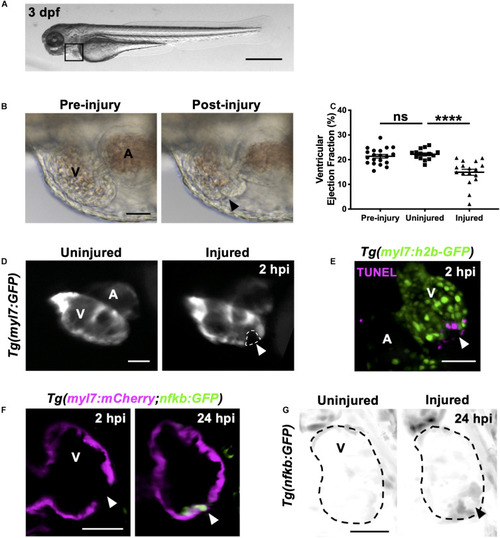
Heart laser injury causes localized cardiomyocyte cell death. |
|
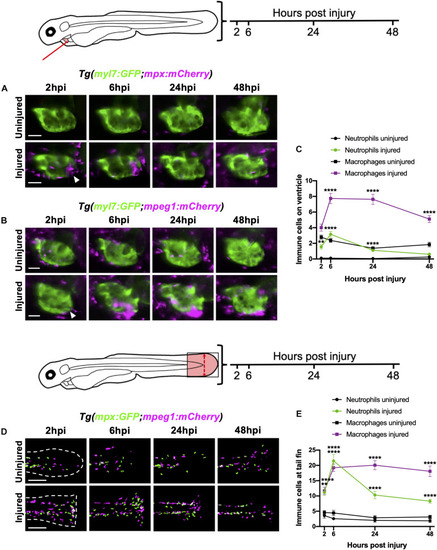
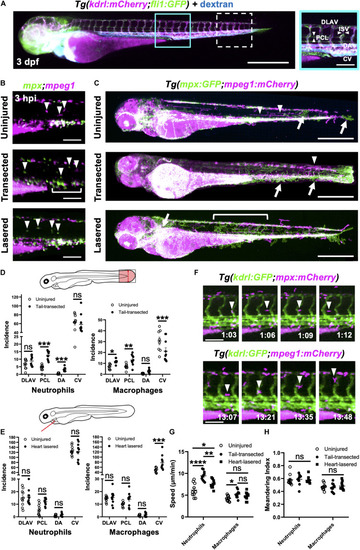
Neutrophils and macrophages display distinct recruitment dynamics during heart injury comparable to tail fin injury. |
|
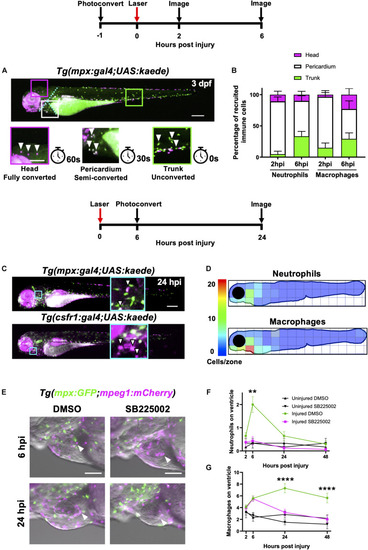
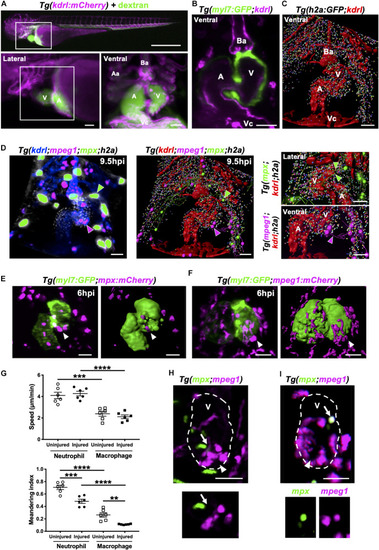
Heart injury recruits local immune cells from the pericardium and head but also cells from distal immune reservoirs. |
|
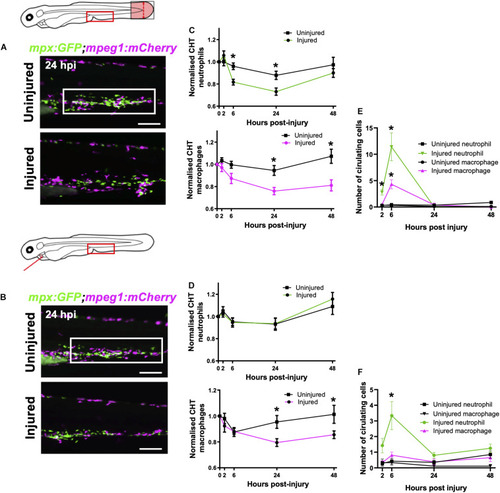
Macrophages are mobilized from the CHT and neutrophils are mobilized into peripheral blood following tail and heart injury. |
|
Neutrophils and macrophages utilize blood and lymphatic vessel surfaces for migration following injury. |
|
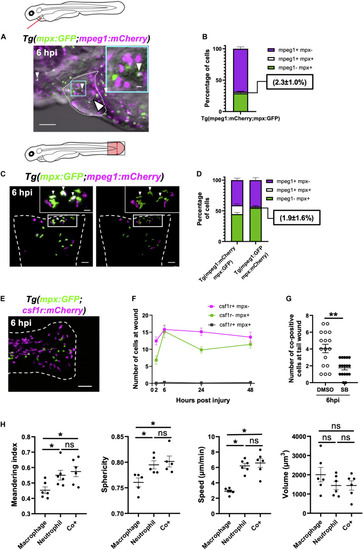
Neutrophils and macrophages migrate onto the ventricle via the pericardium and adopt specific migratory behaviors once on the ventricle following injury. |
|
Immune cells co-positive for |
|
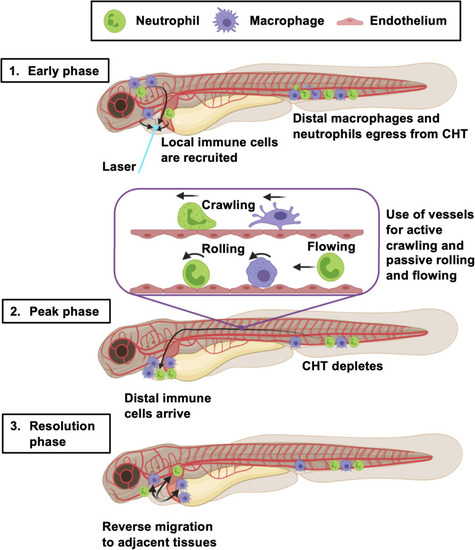
Summary of the larval zebrafish immune response to heart injury. Following laser heart injury in the early phase (0–2 hpi), immune cells resident to nearby pericardial tissues are recruited to the ventricle. At the same time, distal neutrophils and macrophages begin to egress from the CHT into the vascular system. Distal macrophages and neutrophils subsequently utilize blood and lymphatic vessels to migrate to the injured heart, joining locally recruited immune cells during the peak phase (6–24 hpi). The majority of these cells actively crawl along the abluminal surface of vessels, but some also roll and flow along the inside of vessels. The resolution phase is characterized by reverse migration of immune cells to adjacent tissues (> 6 hpi). Created with |