- Title
-
Protein Phosphatase 1 beta Paralogs Encode the Zebrafish Myosin Phosphatase Catalytic Subunit
- Authors
- Jayashankar, V., Nguyen, M.J., Carr, B.W., Zheng, D.C., Rosales, J.B., Rosales, J.B., and Weiser, D.C.
- Source
- Full text @ PLoS One
|
Embryonic Expression of ppp1cba and ppp1cbb during zebrafish development. Detection of ppp1cba and ppp1cbb mRNA was carried out by whole-mount in situ hybridization using gene-specific probes on staged embryos from 256 cells to 24 hpf. Images are lateral views, animal pole at top. (A–L) ppp1cb transcripts are ubiquitously expressed at early developmental stages, 256 cells stage (A and F), sphere stage (B and G), shield stage (C and H), bud stage (D and I) and 24 hpf (K and L). A probe for ppp1cba was used in (A–D and K) while ppp1cbb was used for (F–I and L). Negative control sense probes for ppp1cba and ppp1cbb did not show staining (E and J). Gene specific primers were used to detect ppp1cba and ppp1cbb in various stages of development by RT-PCR. (M) Both genes were expressed maternally and zygotically throughout early development. Amplification of eF1&alpha and total RNA without addition of reverse transcriptase were used as controls. |
|
PP1β paralogs are required for proper zebrafish body axis elongation. (A–I) Lateral views of representative 48 hpf zebrafish embryos injected with (A) uninjected control, (B) 2.5 ng ppp1cba MO, (C) 2.5 ng ppp1cbb MO, (D) a mixture of 0.75 ng ppp1cba MO and 0.75 ng of ppp1cbb MO (2MO), (E) 200 pg of ppp1cba mRNA, (F) 200 pg of ppp1cbb mRNA (G) a partially rescued embryo injected with 100 pg ppp1cba mRNA and 2.5 ng of ppp1cbb MO (H) a partially rescued embryo injected with 100 pg ppp1cbb mRNA and 2.5 ng of ppp1cbb MO (I) a partially rescued embryo injected with 100 pg ppp1cbb mRNA and 0.75 ng of ppp1cbb and 0.75 ppp1cba MO (J) Quantification of the truncated body axis phenotype in morphant and mRNA injected embryos. Each injection was performed multiple times with 50 embryos used to calculate body axis length and reported as % of uninjected clutch mates. Error bars are standard error and a black * indicates a statistically significant difference from control and a # indicates a statistically significant rescue compared to the corresponding morpholino injected embryos. Statistical significance was calculated using a one-factor ANOVA with Tukey post hoc analysis and is defined as p < 0.05. (K) A western blot showing PP1β levels in zebrafish embryo lysates from control and embryos injected with two doses of morpholino cocktail (1.5 ng and 3.0 ng of total MO), individual ppp1cba or ppp1cbb (2.5 ng of either) morpholinos or a rescued embryo injected with 0.75 ng of each morpholino (1.5 ng total) and 100 pg of ppp1cbb mRNA. Tubulin was used as a loading control. |
|
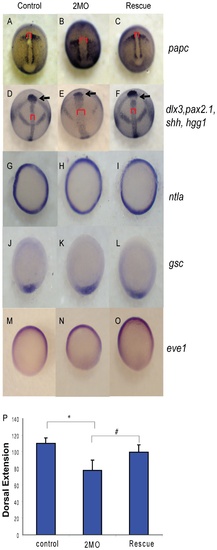
PP1β knockdown blocks convergence and extension but does not alter mesodermal cell fate or dorsal-ventral patterning. Embryos injected with either 1.5 ng of the 2 MO cocktail (0.75 ng ppp1cba MO and 0.75 ng of ppp1cbb MO) or 1.5 ng of the cocktail with 200 pg of ppp1cbb mRNA (rescue) were grown alongside uninjected clutch-mates (control) and staged and fixed for in situ hybridization. Embryos at 90% epiboly were fixed and stained with (A–C) papc (presomitic mesoderm) and imaged from an angle approximately 30 degrees from dorsal to allow visualization of the prechordal plate (marked with an arrow). Embryos at bud stage stained with hgg1 (to mark the prechordal plate), shh (midline), pax2.1 (midbrain-hindbrain boundary) and dlx3 (neural plate) (D–F) and imaged with a view from dorsal. The red bracket marks the width of the notochord and the black arrow points to the prechordal plate. Embryos at 50% epiboly were stained with ntla (G–I) to assay for mesoderm induction, gsc to assay for dorsal cell fates (J–L) and eve1 (M, N, O) for ventral cell fates, all viewed from the animal pole, with dorsal facing down. (P) Quantification of body axis elongation at bud stage for embryos injected with the indicated reagents. Each injection experiment was performed at least 3 times and the morphogenetic measurements were performed on 50 48 hpf and 50 bud stage embryos. Error bars are standard error and a black * indicates a statistically significant difference from control and a # indicates a statistically significant rescue compared to morpholino injected embryos. Statistical significance was calculated using a one-factor ANOVA with Tukey post hoc analysis and is defined as p < 0.05. EXPRESSION / LABELING:
PHENOTYPE:
|
|
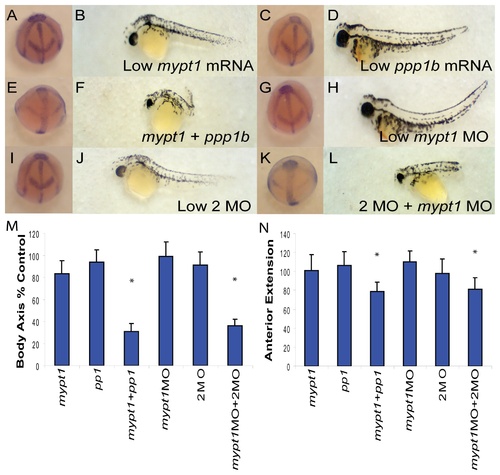
PP1β genes interact genetically with Mypt1. (A–L) Representative embryos at bud stage (A, C, E, G, I, K) and 48 hpf (B, D, F, H, J, L) injected with 15 pg mypt1 (1-300) mRNA with 100 pg of GFP mRNA (A–B), 50 pg each of ppp1cba and ppp1cbb mRNA with 15 pg GFP mRNA (C–D), 15 pg mypt1 mRNA + 50 pg each of ppp1cba and ppp1cbb mRNA (E–F), 0.5 ng of mypt1 MO and 0.25 ng of the control MO (G–H), 0.5 ng control MO and 0.25 ng 2MO (I–J), 0.5 ng mypt1 MO and 0.25 ng 2MO (0.125 ng ppp1cba MO and 0.125 ng of ppp1cbb MO) (K–L). Quantification of the angle of body axis extension at bud stage (M) and 48 hpf (N). Error bars are standard error and a * indicates a statistically significant difference from control and a # indicates a statistically significant rescue compared to morpholino injected embryos. For statistical analysis 50 embryos were analyzed for each condition. Statistical significance was calculated using a one-factor ANOVA with Tukey post hoc analysis and is defined as p < 0.05. |
|
PP1β paralogs are required for cell shape changes required for convergent extension. A representative field of presomitic and notochordal mesoderm at the bud stage in control (A), 2MO (0.75 ng ppp1cba MO and 0.75 ng of ppp1cbb MO) embryos (B) or rescued embryos (C). The cell polarity of presomitic (D) and notochordal (E) mesodermal cells was determined by calculating the length width ratio (y-axis). The crossed bars indicate cells with the long axis length and the short axis width. The embryos are arranged such that dorsal is down and anterior is to the right. Arrows indicate cells that are actively producing bleb-like protrusions. Error bars are standard error and a * indicates a statistically significant difference from control. All calculations were made on 75 cells from 3–5 separate embryos. Statistical significance was calculated using a one-factor ANOVA with Tukey post hoc analysis and is defined as p < 0.05. PHENOTYPE:
|
|
The ppp1cba/ppp1cbb knockdown phenotype is independent of p53. (A–C) Lateral views of representative 48 hpf zebrafish embryos injected with (A) 4 ng p53 MO, (B) 0.75 ng ppp1cba MO, 0.75 ng of ppp1cbb MO (2MO) and 4 ng p53 MO (C) a partially rescued embryo injected with 100 pg ppp1cbb mRNA, 0.75 ng of ppp1cbb, 0.75 ppp1cba MO and 4 ng p53 MO. (D) In addition, control embryos were injected with 4 ng control MO; 2MO and 4 ng control or 2MO, 100 pg ppp1cbb mRNA and 4 ng of control MO (D) Quantification of the truncated body axis phenotype in morphant and mRNA injected embryos. Each injection was performed multiple times with 25 embryos used to calculate body axis length and reported as % of uninjected clutch mates. Error bars are standard error, a black * indicates a statistically significant difference from control, # indicates a statistically significant rescue and a NS indicates two data sets that are not significantly different. Statistical significance was calculated using a one-factor ANOVA with Tukey post hoc analysis and is defined as p < 0.05. |
|
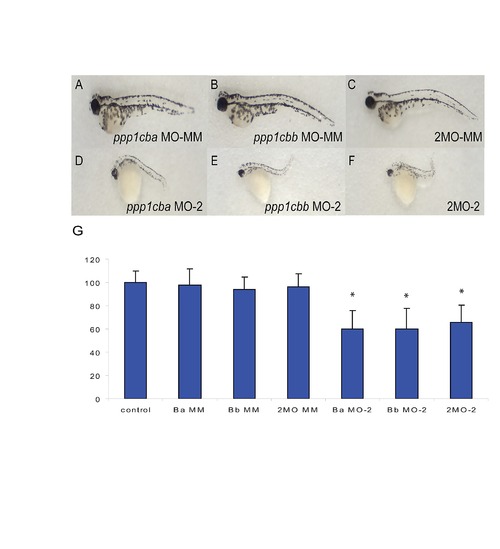
Body axis elongation defects are induced by alternative ppp1cba and ppp1cbb morpholinos but not mismatched controls. (A–C) Lateral views of representative 48 hpf zebrafish embryos injected with (A) 2.5 ng mismatch ppp1cba MO, (B) 2.5 ng mismatch ppp1cbb MO, (C) a mixture of 0.75 ng mismatch ppp1cba MO and 0.75 ng of mismatch ppp1cbb MO (2MO-MM), (D) 5.0 ng ppp1cba MO-2, (E) 5.0 ng ppp1cbb MO-2, (F) a mixture of 1.5 ng ppp1cba MO and 1.5 ng of ppp1cbb MO (2MO-2). (G) Quantification of the truncated body axis phenotype in morphant and control embryos. Each injection was performed multiple times with 25 embryos used to calculate body axis length and reported as % of uninjected clutch mates. Error bars are standard error and a black * indicates a statistically significant difference from control. Statistical significance was calculated using a one-factor ANOVA with Tukey post hoc analysis and is defined as p < 0.05. |