- Title
-
The hypothalamic neuropeptide oxytocin is required for formation of the neurovascular interface of the pituitary
- Authors
- Gutnick, A., Blechman, J., Kaslin, J., Herwig, L., Belting, H.G., Affolter, M., Bonkowsky, J.L., and Levkowitz, G.
- Source
- Full text @ Dev. Cell
|
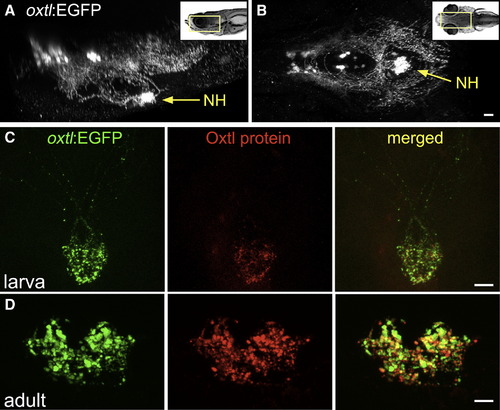
Oxytocinergic Reporter Transgene Tags HN Projections (A and B) 3D reconstruction of the hypothalamic-neurohypophseal system of an optically sectioned 9-day-old zebrafish embryo carrying the oxtl:EGFP transgene (anterior to the left; A, lateral view; B, ventral view). Axonal projections to the neurohypophysis (NH) are highly visible in oxtl:EGFP transgenic animals due to extensive arborization of NH nerve termini. (C and D) Immunohistochemical analysis of Oxytocin protein in oxtl:EGFP transgenic zebrafish showing colocalization of EGFP+ and Oxytocin+ nerve termini in the neurohypophysis of either 6-day-old larva (C, anterior up) or coronally sectioned adult (D); 14-day-old). Scale bars, 20 μm. See also Figure S1 and Movie S1. EXPRESSION / LABELING:
|
|
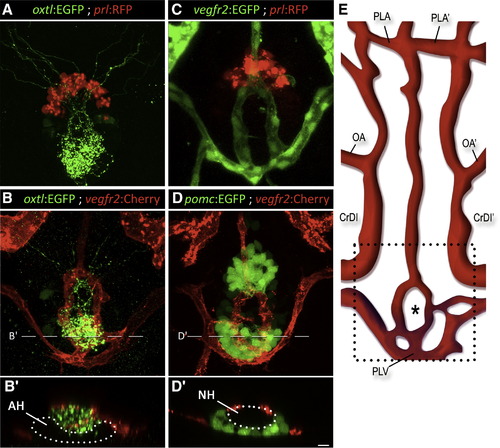
Axovasal Interactions in the Zebrafish Pituitary (A–D) Three-dimensional reconstructions of the hypophysis of 9-day-old zebrafish embryos carrying pairs of transgenic fluorescent markers of different hypophyseal components. (A) The oxtl:EGFP transgene (green) marks oxytocinergic axons terminating at the neurohypophysis, just dorsal and posterior to the adenohypophyseal prolactin-producing cells marked with prl:RFP (red). (B) Double transgenic animals expressing oxtl:EGFP (green) together with the vascular endothelial marker vegfr2:Cherry (red) reveal a distinct, previously unidentified structure of the zebrafish hypophyseal vasculature. (C and D) Hypophyseal vasculature, visualized by either vegfr2:EGFP or vegfr2:Cherry, together with the adenohypophyseal markers prl:RFP and pomc:EGFP. The ring-like vascular structure resided dorsal to the adenohypophysis. (B2) and (D2) show optical Z-slices that demonstrate the position of the NH in an indentation of the adenohypophysis. (E) Schematic map of ventral head vasculature of the zebrafish larvae, including the location of the hypophyseal vasculature. The asterisk marks the location of the neurohypophysis. AH, adenohypophysis; CrDI, cranial division of the internal carotid artery; NH, neurohypophysis; OA, optic artery; PLA, palatocerebral artery; PLV, palatocerebral vein; pomc, proopiomelanocortin; prl, prolactin; vegfr, vascular endothelial growth factor receptor. Scale bar, 20 μm. See also mmc3VIDEO, mmc4VIDEO and mmc5VIDEO. EXPRESSION / LABELING:
|
|
3D Structure of the Hypophyseal Neurovascular Interface (A and B) Identification of hypophyseal arteries and veins by real-time analysis of hypophyseal blood flow. (A) Confocal time-lapsed images (t1–t10) of a single gata1+ blood cell (arrowhead) traveling from the hypophyseal artery into the neurohypophysis and its outflow through the hypophyseal veins. Superposition of all time points and the direction of blood flow (red arrow) are shown in the right panel. Adjacent Prl-RFP+ adenohypophyseal cells (arrow) served as a positional landmark. (B) Schematic map of ventral head vasculature of the zebrafish larvae, including the newly annotated hypophyseal artery (red) and veins (blue). AH, adenohypophysis, CrDI, cranial division of the internal carotid artery; HyA, hypophyseal artery; HyV, hypophyseal veins; OA, optic artery; PLA, palatocerebral artery. (C) Schematic representation of the 3D structure of the zebrafish pituitary depicting the various cellular components of the adeno- and neurohypophysis. A longitudinal section of this structure is shown on the right. See also Figure S2 and Movie S5. EXPRESSION / LABELING:
|
|
Morphogenesis of the Neurohypophysis (A–C) Dynamics of hypophyseal innervation. Images of transgenic otpb:caax-EGFP embryos reveal that innervation of the hypophysis by neurons of the neurosecretory preoptic nucleus (NPO) occurs as early as 36 hr postfertilization and becomes well established by 54 hr. Scale bar, 20 μm. (D–F) Dynamics of neurovascular interface. Images of double transgenic otpb:caax-EGFP;fli1:dsRed embryos at different developmental stages showing innervation of the neurohypophysis prior to hypophyseal vessel formation. (G) Dynamics of hypophyseal vascularization. Time-lapsed (48–72 hr) confocal microscopy was used to track vascular endothelial cells in the ventral diencephalon of vegfr2:Cherry embryos. The upper panels display snapshots of critical stages in the formation of the hypophyseal loop-like structure. A schematic representation of the corresponding images with arterial (red) and venous (blue) color-coding is shown at the bottom. CrDI, cranial division of the internal carotid artery; HyA, hypophyseal artery; HyV, hypophyseal veins; OA, optic artery; PHS, primary head sinus; PLA, palatocerebral artery. The asterisk (*) demarcates the location of the neurohypophysis. See also Movie S6. EXPRESSION / LABELING:
|
|
Abnormal Development of Pituitary Vasculature in the Absence of HN Innervation (A and B) Immunostaining of oxytocin (Oxtl) in wild-type (A) and the endothelial-deficient mutant cloche (- / -) (B) mutants, which lack all blood vessels. (C–L) Genetic cell ablation of Otp+ neurons of the neurosecretory preoptic nucleus (NPO). (C) Quantitative analysis of the vascular phenotype caused by HN neuronal ablation (see text). Using the Gal4/UAS system (otpb:Gal4;UAS:NTR-Cherry) to drive expression of a Nitroreductase-Cherry (NTR-Cherry) fusion protein only in Otp+ neurons make these cells and their projections visible (red) and susceptible to the prodrug metronidazole. This transgenic fish was crossed to the vascular reporter line, vegfr2:EGFP, to generate a triple transgenic line (D–F and J–L). Double transgenic line (otpb:Gal4;vegfr2:EGFP, G–I) expressing vascular EGFP but not the NTR-Cherry protein served as a control for nonspecific drug toxicity. The presence of Cherry+ neurons (D, G, J), EGFP+ blood vessels (E, H, and K) and oxytocin immunoreactivity (F, I, and L) were monitored in three experimental conditions; the respective genotypes as well as the application of the metronidazole killing agent are indicated below. Filled and empty arrowheads indicate normal and abnormal blood vessels, respectively. See also Figure S3. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Oxytocin and Its Receptor Are Necessary for Pituitary Vascularization (A–D) High-resolution confocal images of the head vasculature of 3-day-old embryos following targeted knockdown of the gene encoding for oxytocin (oxtl-MO) or oxytocin receptor (oxtlr-MO) using antisense morpholino oligonucliotides, leads to severe impairment of neurohypophyseal vasculature, but does not impede development of neighboring vessels. A schematic representation of the corresponding phenotype with arterial (red) and venous (blue) color-coding is shown in (A2)–(C2). (D and E) By crossing nuclear- and membrane-tagged fluorescent vascular reporter lines, we are able to count the number of vascular endothelial cells in each vessel. A dot-plot histogram showing the x,y positions of EGFP+;Cherry+ endothelial cells nuclei of the hypophyseal vessels as well as the adjacent internal carotid arteries (CrDI, between the PICA and the PLA) is shown in (D2) and (E2). Individual cells belonging to the hypophyseal vessels (red) and CrDI (gray) are color coded. The oxtlr-MO-injected embryos (E) display a markedly reduced number of endothelial cells in the hypophyseal vessel but no change in the nearby internal carotid artery. (F) Bar histogram showing EGFP+;Cherry+ cell counts in the internal carotid arteries and hypophyseal vessels in control and following oxtlr knockdown. *p < 0.01, n = 14. (G) RT-PCR analysis of vegfr2:Cherry-positive and -negative FACS sorted endothelial cells demonstrating vascular expression of the oxytocin receptor (oxtlr). CaDI, caudal division of internal carotid; CrDI, cranial division of the internal carotid artery; MO, morpholino antisense oligonucleotide; OA, optic artery; PICA, primitive internal carotid artery; PLA, palatocerebral artery; PLV, palatocerebral vein. The asterisk (*) demarcates the location of the neurohypophysis. See also Figure S4. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Cell-Autonomous Effect of the Oxytocin Receptor on the Pituitary Vasculature (A–C) Genetic complementation of oxytocin receptor in the vascular endothelia of oxtlr-deficient embryos. Double transgenic embryos expressing an endothelial Gal4 driver (fli1:Gal4) and a fluorescent reporter protein (UAS:Kaede; green) were injected with transposon-based transgenic vector containing a multicistronic gene expression cassette (UAS:oxtlr-2A-EGFP) allowing simultaneous mosaic coexpression of the oxytocin receptor and EGFP in discrete vascular clones (red). These clones were detected by immunostaining with an anti-EGFP antibody that does not react with the Kaede protein followed by a secondary cy3-conjugated antibody. The asterisk (*) demarcates the location of the neurohypophysis. (D) Schematic model describing the hypothalamohypophyseal neurovascular connection by local secretion of oxytocin (see text). CaDI, caudal division of internal carotid; CrDI, cranial division of the internal carotid artery; OA, optic artery; PHS, primary head sinus; PLA, palatocerebral artery. |
|
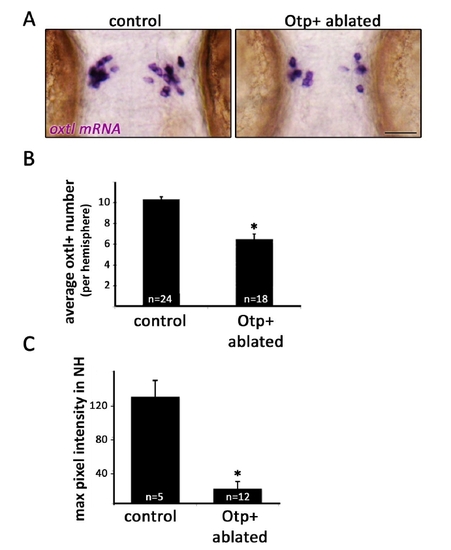
Figure S3 (related to Figure 5). Genetic cell ablation of Otp+ cells of the neurosecretory preoptic nucleus (NPO). A,Micrographs of 3-day old embryos that underwent Otp+ cell ablation and their unablated siblings (see ‘RESULTS’ section). Embryos were subjected to in-situ hybridization with oxtl mRNA probe. Scale bar, 50μm. B, Bar histogram depicting oxtl+ cell counts in control and Otpb+ cell ablated embryos. *p<0.001. C, Oxtl protein was stained by immunofluorescence in 3-day old embryos that underwent Otp+ cell ablation and in their unablated siblings. Maximal pixel intensity (between 0-255) was recorded from confocal images of the NH region of each embryo and normalized for background pixel intensity. Oxtl staining was found to be reduced by ~85% in embryos that had undergone Otpb+ ablation. *p<0.001. |
|
Figure S4 (related to Figure 6). Effects of oxtl and oxtlr gene knockdowns A, Scheme depicting oxtl gene structure indicating the binding site of a splice blocking antisense oligonucleotide (oxtl-MO). B, Reduction of Oxytocin (Oxtl) protein expression by oxtl-directed antisense oligonucleotide (oxtl-MO). Anti-oxytocin immunostaining of the neurohypophysis of 3-day old transgenic oxtl:EGFP embryos. Injection of oxtl-MO causes a marked reduction in Oxtl but not EGFP protein levels. C, Scheme depicting oxtlr gene structure indicating the binding site of a splice blocking antisense oligonucleotide (oxtlr-MO) as well as the PCR primers used to amplify the respective exons (Ex) of the various mRNA products. A table summarizing the expected mRNA sizes in the control and followed oxtlr knockdown is shown at the bottom. D, Gel electrophoresis analysis of PCR-amplified mRNA species in control and in embryos injected with oxtlr-MO. Primer pairs used to amplify the respective exons (Ex) are indicated at the top. Amplification of exon 1 (Ex1F/Ex1R) was used as control to demonstrate untargeted constitutive exon, which was not affected by the knockdown reagent. E, Scatterplot representation of the fluorescence profiles of cell suspensions from dissociated 3-day old vegfr2:mCherry embryos (right) and their non-transgenic siblings (left), prior to FACS sorting. P1 is the proportion of cells that exhibit high red fluorescence (defined as ‘mCherry+’ cells, ~1% of the total population). P4 is the proportion of cells that exhibit no fluorescence (defined as ‘mCherry-’ cells, ~80% of the total population). |
Reprinted from Developmental Cell, 21(4), Gutnick, A., Blechman, J., Kaslin, J., Herwig, L., Belting, H.G., Affolter, M., Bonkowsky, J.L., and Levkowitz, G., The hypothalamic neuropeptide oxytocin is required for formation of the neurovascular interface of the pituitary, 642-654, Copyright (2011) with permission from Elsevier. Full text @ Dev. Cell