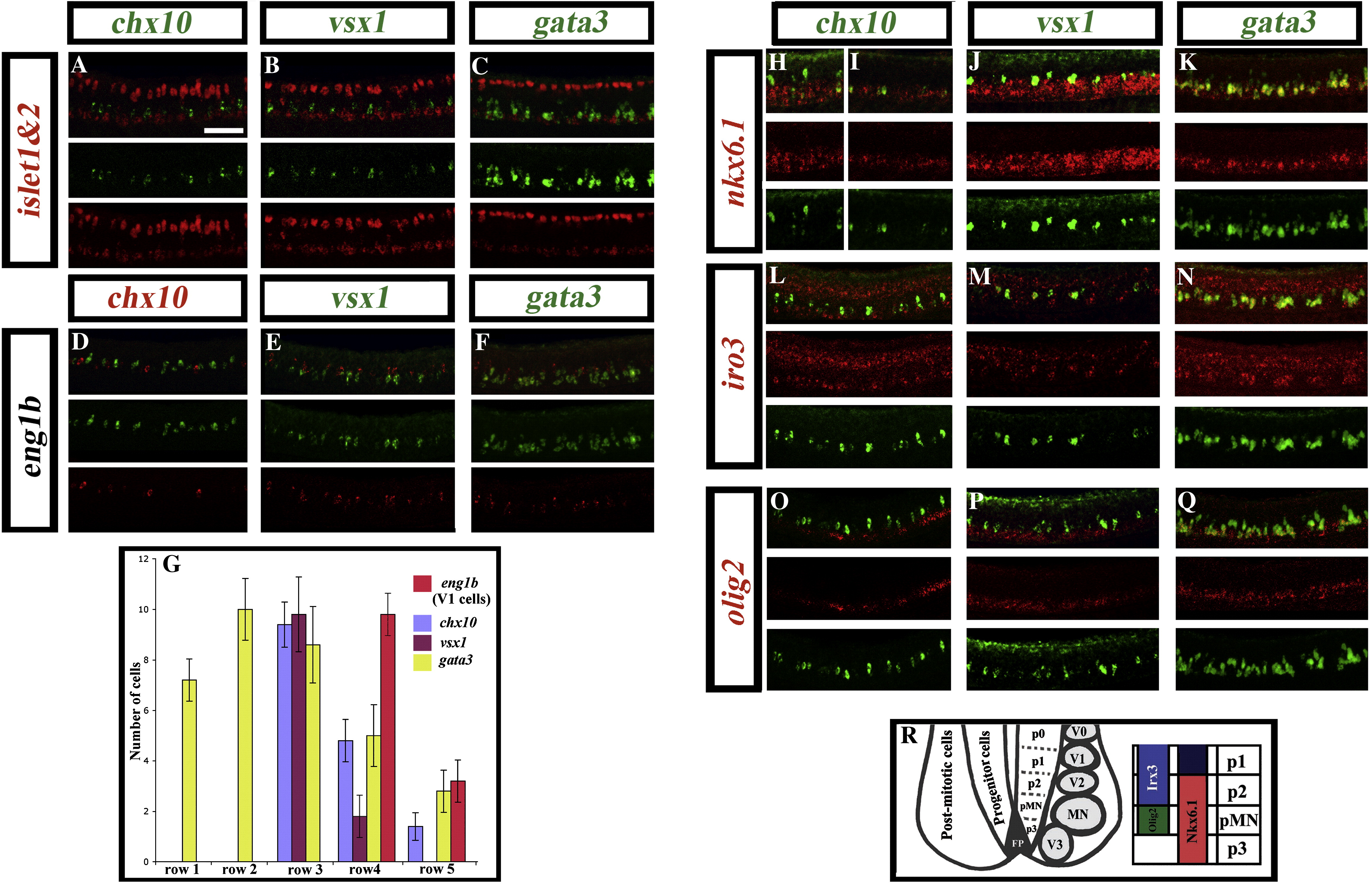
Fig. 2
Fig. 2 Expression of transcription factors in the V2 domain. Lateral views of double in situ hybridisation in wild-type trunks at 24 h. islet1 and islet2 (probes mixed) are shown in red (A–C); eng1b is shown in green (D) and red (E and F); chx10 is shown in green (A, H, I, L and O) and red (D); vsx1 is shown in green (B, E, J, M and P); gata3 is shown in green (C, F, K, N and Q); nkx6.1, iro3 and olig2 are all shown in red (H–Q). Panel H shows expression in the anterior of the embryo and panel I shows expression in the posterior of the embryo. Scale bar = 50 μm. (G) Graph showing the number of cells in rows 1–5 at 24 h expressing V2 markers gata3, chx10 or vsx1 or V1 marker eng1b. None of these genes are expressed dorsal to row 5 at this stage. All cell counts are from a 5 somite length of spinal cord adjacent to somites 6–10. Values shown are the mean from 5 different embryos. Error bars indicate standard deviations. (R) Schematic showing the dorsal–ventral location of different ventral post-mitotic interneuron domains (V0–V3), the motoneuron domain (MN) and transcription factors that demarcate the p2 progenitor domain.
Reprinted from Developmental Biology, 322(2), Batista, M.F., Jacobstein, J., and Lewis, K.E., Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons, 263-275, Copyright (2008) with permission from Elsevier. Full text @ Dev. Biol.