Figure 5—figure supplement 3.
- ID
- ZDB-FIG-201012-100
- Publication
- Heger et al., 2020 - The genetic factors of bilaterian evolution
- Other Figures
-
- Figure 1
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 2.
- Figure 1—figure supplement 3.
- Figure 2
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 2.
- Figure 2—figure supplement 3.
- Figure 2—figure supplement 4.
- Figure 3.
- Figure 4
- Figure 4—figure supplement 1.
- Figure 5
- Figure 5—figure supplement 1.
- Figure 5—figure supplement 2.
- Figure 5—figure supplement 3.
- Figure 5—figure supplement 4.
- Figure 6
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 2.
- Figure 6—figure supplement 3.
- Figure 7
- Figure 7—figure supplement 1.
- Figure 8.
- All Figure Page
- Back to All Figure Page
|
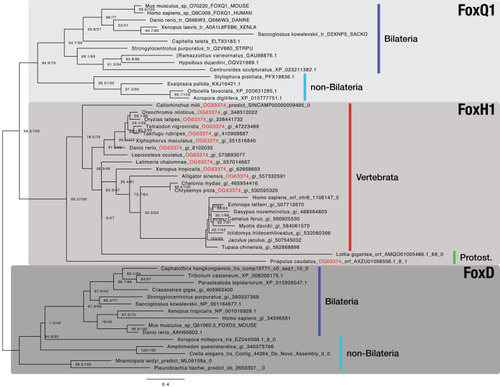
Maximum likelihood phylogeny of selected metazoan Fox genes. The multiple sequence alignment consists of 52 sequences aligned over 315 positions (proportion of gaps and undetermined characters: 25.07%). It is generated from OG_36001 (FoxH1), OG_63374 (RBH with OG_36001; orthogroup ID labeled in red), and representative sequences of OG_3972 (FoxD4 as outgroup; third-best hit of OG_36001 in HMM-HMM searches, see |