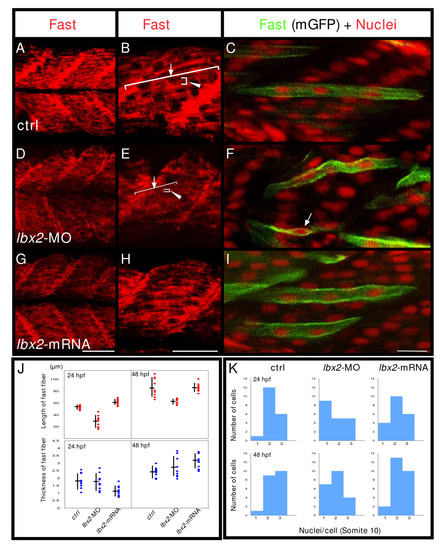
Fast muscle fibers are malformed in the absence of Lbx2. A-C: Control embryos. D-F: lbx2-MO injected embryos. G-I: lbx2 mRNA injected embryos. (A, B, D, E, G, H, J) Embryos labeled with the fast muscle marker, EB165 (red). (B, E, H) Higher magnification views. Rostral caudal extension of fast fibers is severely affected (B, E, arrow). J: Statistical analyses of mean filament rostral-caudal length (B, E, arrows) or dorsal ventral thickness (B, E, arrowhead). Analysis by ANOVA demonstrates significant differences in length (P < 0.01). (C, F, I) Embryos were mosaically labeled with membrane localized GFP (mGFP) by injection of DNA and labeled for nuclei (red, propidium iodide). mGFP expressing cells in the fast muscle domain at the level of somite 10 were randomly selected and the number of their nuclei counted. K: Fast muscle cells are multinucleate by 48 hpf in both control and lbx2 mRNA injected embryos, whereas unfused fast muscle cells are observed in lbx2-MO injected embryo (F, arrow). Analysis demonstrates significant differences among the three groups at 48 hpf (Kruskal-Wallis test, p = 0.0121). No apparent differences can be detected between controls and embryos injected with lbx2-MO at 24 hpf (Kruskal-Wallis test, p = 0.1509). (A-I) Lateral views, rostral toward the left, dorsal toward the top. Scale bars: (A, B, D, E, G, H) 50 μm, (C, F, I) 20 μm.
|

