- Title
-
Biocompatible exosomes derived from Pinctada martensii mucus for therapeutic melanin regulation via α-MSH/NF-κB/MITF pathway
- Authors
- Mo, D., Zheng, W., Gao, Z., Ma, K., Yang, K., Zeng, T., Qin, C., Luo, Y., Zheng, L., Xu, S.
- Source
- Full text @ Regen Biomater
|
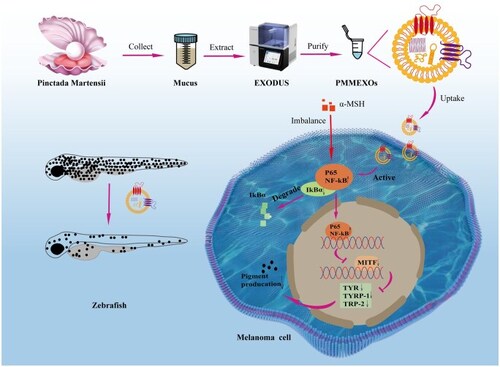
Exosomes derived from |
|
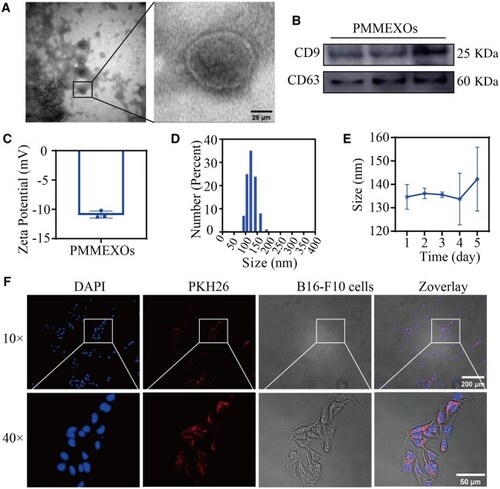
Characterization of PMMEXOs. ( |
|
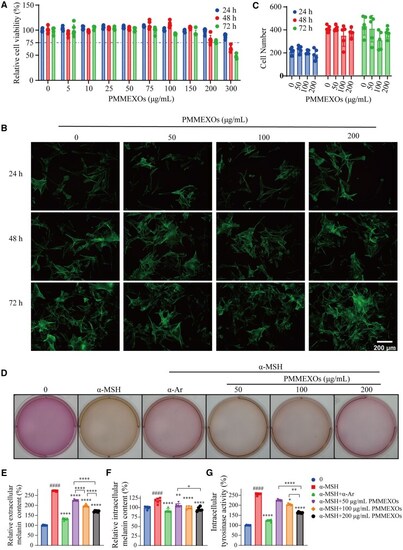
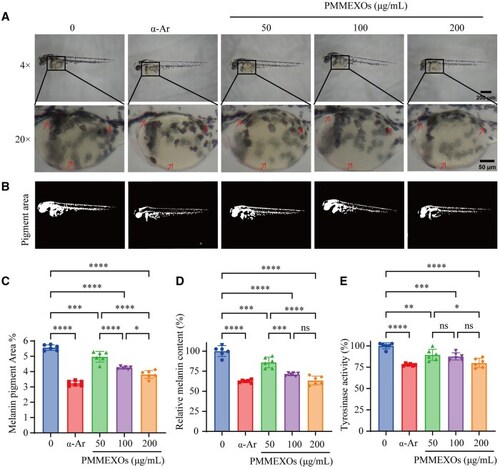
Effects of PMMEXOs on melanin secretion and tyrosinase activity in B16F10 melanoma cells. ( |
|
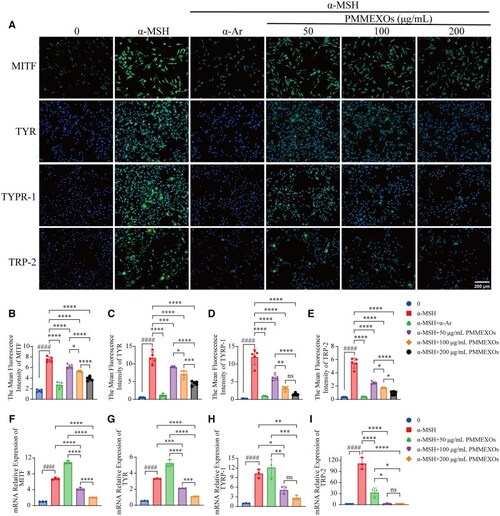
PMMEXOs inhibits the expression of melanin-related proteins and genes. ( |
|
|
|
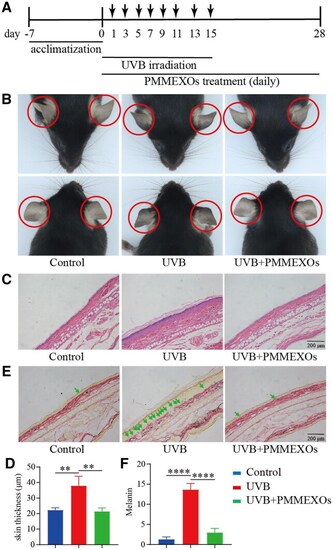
The effect of PMMEXOs on excessive pigmentation of the ear skin in C57BL/6 mice induced by UVB irradiation. ( |
|
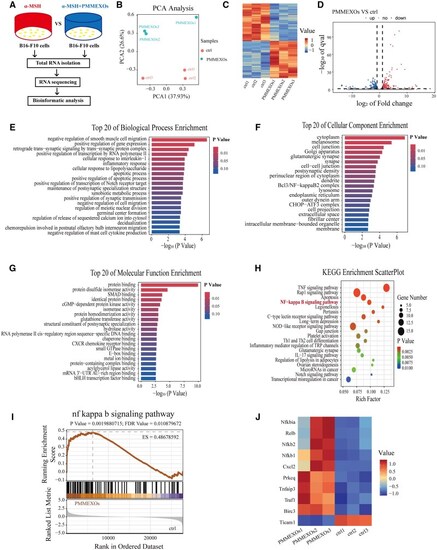
Analysis of RNA sequencing results and enrichment of differentially expressed genes. ( |
|
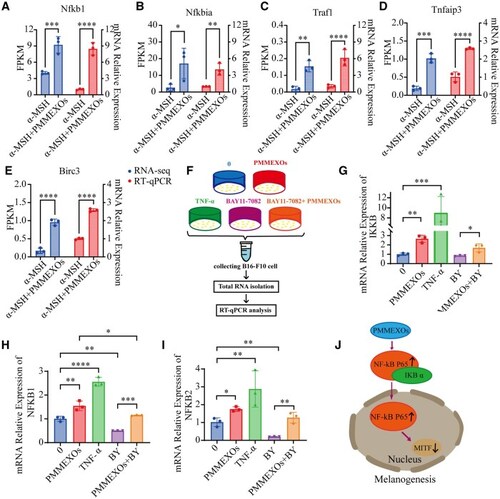
RT-qPCR validated transcriptome sequencing data. Confirmation of transcriptome sequencing data by RT-PCR, including Nfkb1 ( |
|
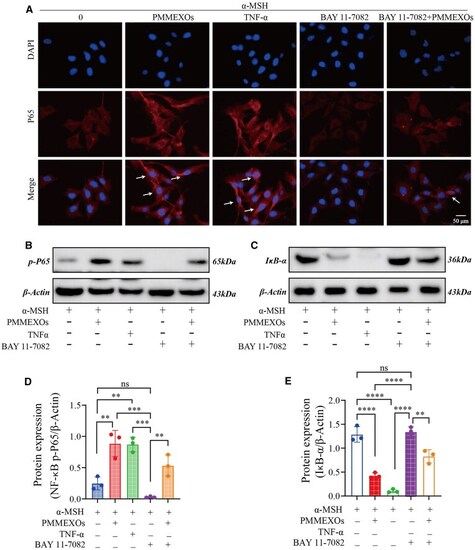
PMMEXOs reduces melanin production by activating the NF-κB signaling pathway. ( |
|
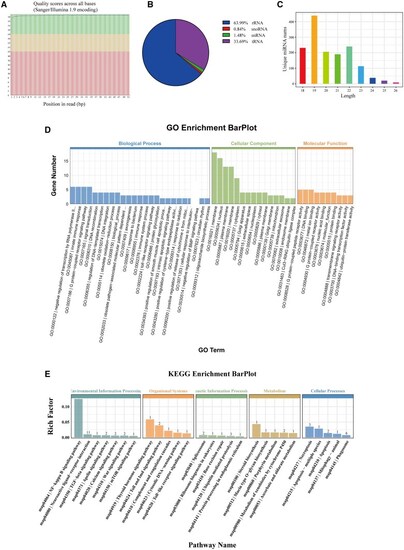
miRNA-Seq analysis of PMMEXOs. ( |