- Title
-
Creb3l3 deficiency promotes intestinal lipid accumulation and alters ApoB-containing lipoprotein kinetics
- Authors
- Sweeney, D.W., Shen, M.C., Farber, S.A.
- Source
- Full text @ J. Lipid Res.
|
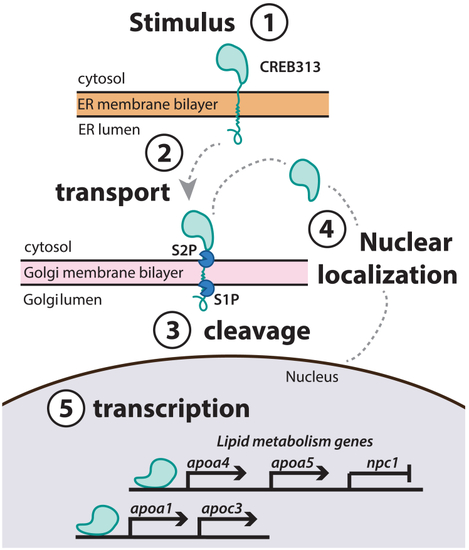
Process of CREB3L3 nuclear localization. 1: In the inactive state, CREB3L3 is anchored to the membrane of the endoplasmic reticulum (ER). 2: An unknown signal induces the release of the tethered protein from the ER membrane, and CREB3L3 is transported to the Golgi. 3: Proteases (S1P, S2P) cleave the transmembrane domain of the protein in the Golgi. 4: Upon cleavage, the DNA-binding domain of the protein can be trafficked to the nucleus. 5: CREB3L3 regulates expression of key lipid metabolism and apoprotein genes by binding to promoter regions and altering transcriptional processes in the nucleus. CREB3L3, cAMP-responsive element-binding protein 3–like 3. |
|
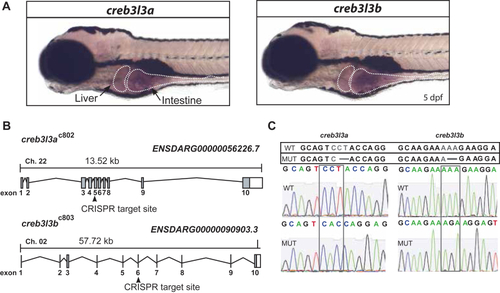
Generation of |
|
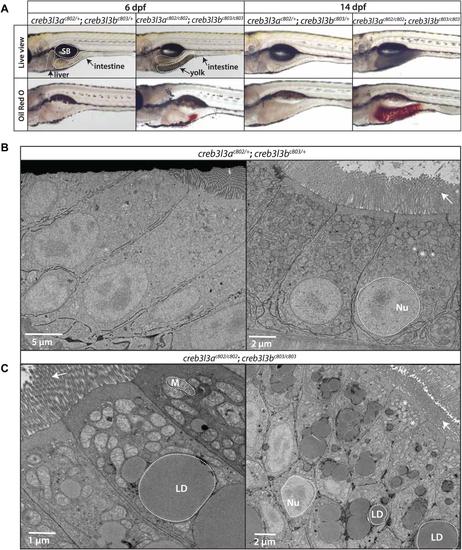
Accumulation of cytoplasmic lipid droplets in the enterocyte of PHENOTYPE:
|
|
|
|
|
|
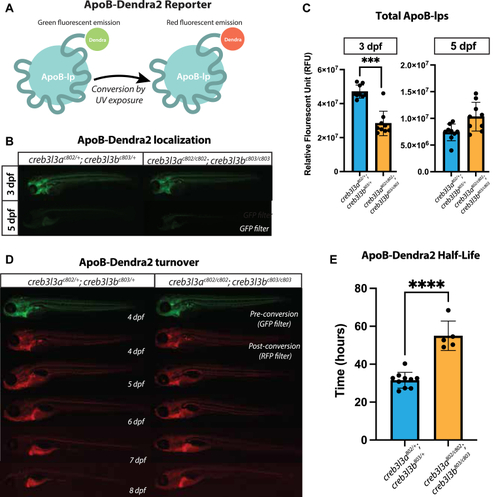
ApoB-Dendra2 reporter localization in |
|
The loss of Creb3l3 may affect lipid secretion from the enterocyte. A: Fluorescent signal of BODIPY-C12 within intestinal homogenate over 24 h post gavage shows no significant difference between |
|
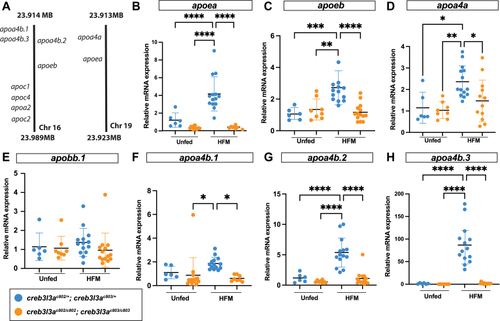
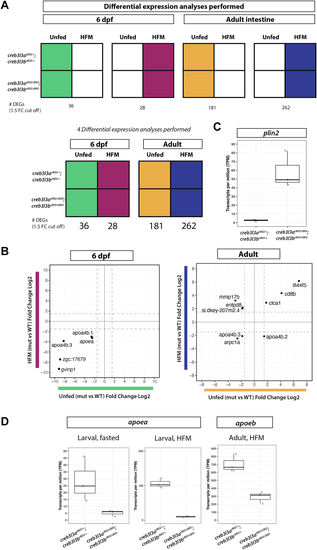
RNA sequencing analysis of |
|
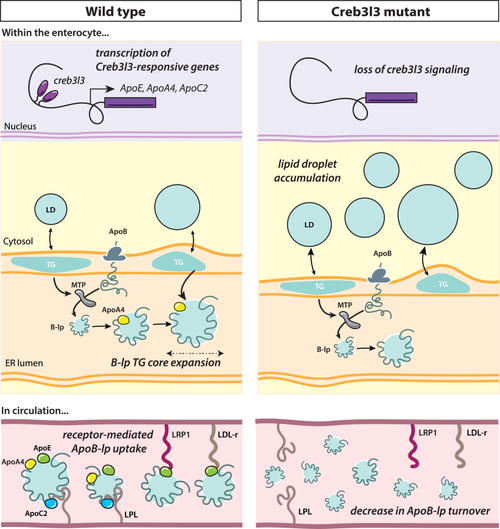
Summary of results and hypothetical model. Creb3l3 signaling activates Creb3l3-responsive genes such as ApoC2, ApoE, and ApoA4. We found that loss of Creb3l3 results in an increase of cytoplasmic lipid droplets in the enterocytes likely due to the loss of ApoA4. It is speculated that ApoA4 is responsible for expanding the size of the ApoB-lp through the addition of triglyceride potentially in poorly understood post ER steps. If enterocytes are unable to enlarge particles, the enterocyte may be directing excess lipid to cytoplasmic lipid droplets for storage. In circulation, ApoE facilitates the uptake of B-lp by peripheral tissues. In our study, we hypothesize that the increase of ApoB-lp half-life, or decrease of ApoB-lp turnover, is a direct result of the loss of ApoE from the ApoB-lp particle. ApoB-lp, apolipoprotein-B–containing lipoprotein; B-lp, ApoB-containing lipoprotein; Creb3l3, cAMP-responsive element-binding protein 3–like 3; ER, endoplasmic reticulum; LDL-r and LRP1, members low-density lipoprotein receptor family; LPL, lipoprotein lipase; MTP, microsomal triglyceride transfer protein; TG, triglyceride. |