- Title
-
KDM6A Deficiency Induces Myeloid Bias and Promotes CMML-Like Disease Through JAK/STAT3 Activation by Repressing SOCS3
- Authors
- Chen, H., Wang, S., Dong, R., Yu, P., Li, T., Hu, L., Wang, M., Qian, Z., Zhou, H., Yue, X., Wang, L., Xiao, H.
- Source
- Full text @ Adv Sci (Weinh)
|
|
|
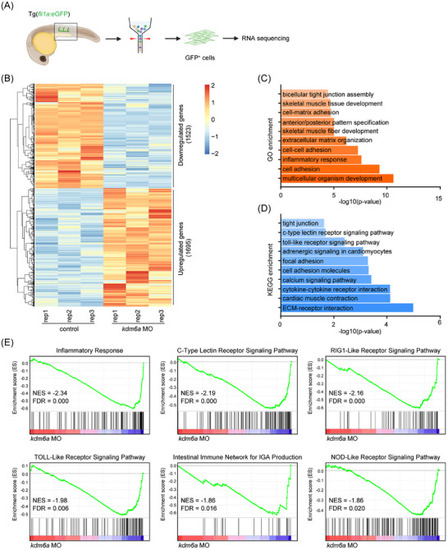
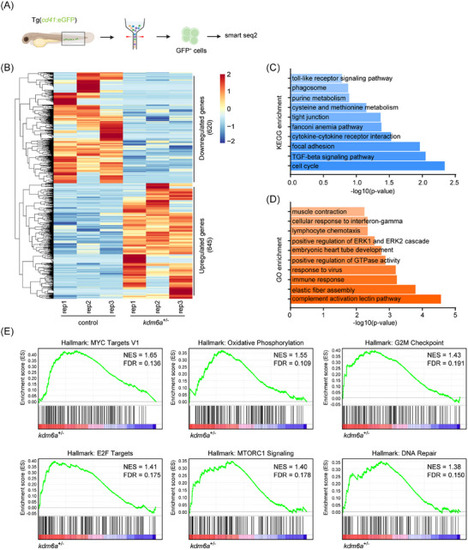
Loss of Kdm6a deregulates inflammatory gene expression in ECs. A) Flowchart of sorting and RNA sequencing. B) Heatmap of differentially expressed genes in endothelial cells from |
|
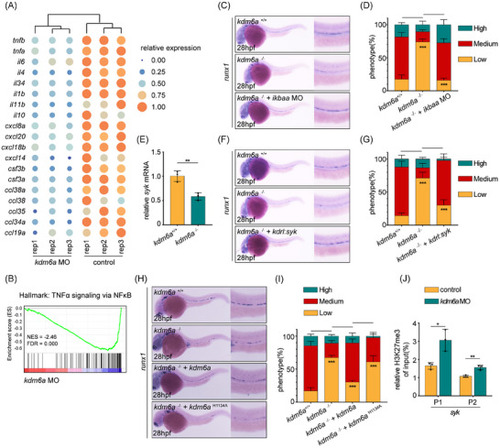
Kdm6a regulates HSPC production via Syk‐associated inflammatory response in a H3K27me3‐dependent manner. A) Heatmap showing differential expression of inflammatory‐associated genes. B) Enrichment plot of the TNFα signaling via NF‐κB between differentially regulated genes in |
|
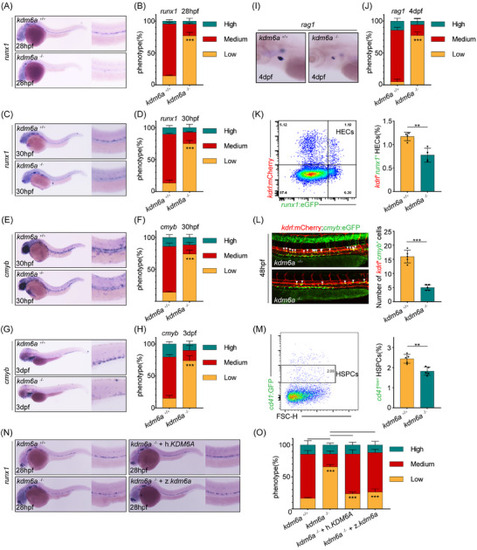
Haploinsufficiency of Kdm6a has skewed myelopoiesis in embryonic and adult zebrafish. A,B) WISH for |
|
Haploinsufficiency of Kdm6a dysregulates aging related gene expression in HSPCs. A) Flowchart of sorting and RNA sequencing. B) Heatmap of differentially expressed genes in HSPCs from |
|
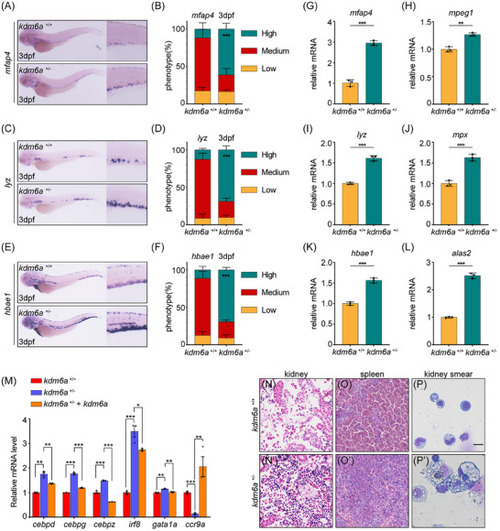
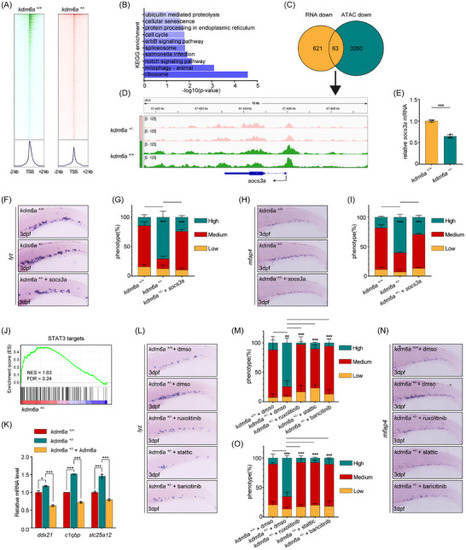
Haploinsufficiency of Kdm6a promotes myeloid‐biased hematopoiesis through repressing |
|
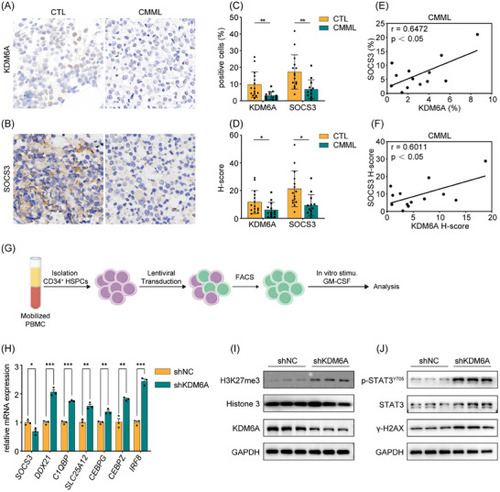
KDM6A/SOCS3/p‐STAT3 pathway is conserved in human HSPCs. A) Immunohistochemical staining of KDM6A on BM biopsies. B) Immunohistochemical staining of SOCS3 on BM biopsies. C) The percentage of KDM6A+ and SOCS3+cells in immunohistochemical staining. D) The H‐score of KDM6A+ and SOCS3+ cells immunohistochemical staining. E) Correlation analysis of the percentage of KDM6A+ (C, left) and SOCS3+ (C, right) cells in CMML specimens. F) Correlation analysis of the H‐score of KDM6A+ (D, left) and SOCS3+ (D, right) cells in CMML specimens. G) Schematic representation of human HSPC enrichment, lentiviral infection, and in vitro stimulation experiments conducted in this study. H) qPCR analysis of genes expression including |