- Title
-
Zbtb48 is a regulator of Mtfp1 expression in zebrafish
- Authors
- Goh, S.Y.C., Fradera-Sola, A., Wittkopp, N., Şerifoğlu, N., Godinho Ferreira, M., Ketting, R.F., Butter, F.
- Source
- Full text @ Commun Biol
|
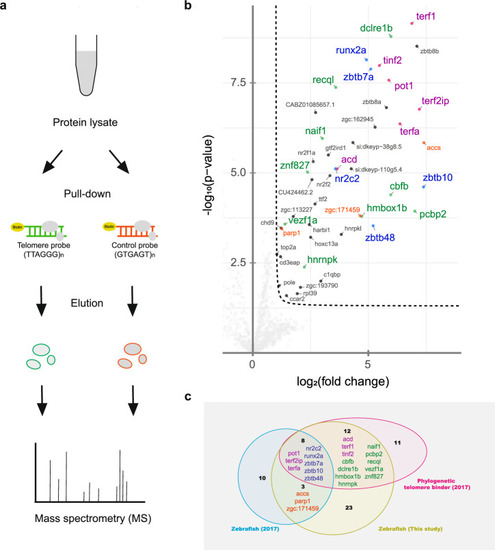
Telomere pull-down with the zebrafish cell line. |
|
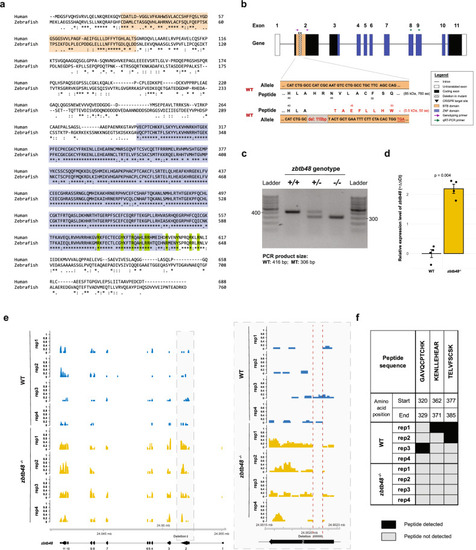
Generation and validation of the |
|
|
|
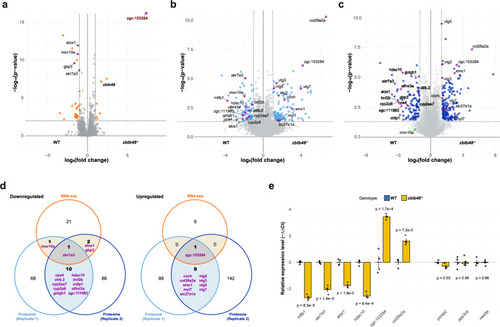
Transcriptomic and proteomic profiling of |
|
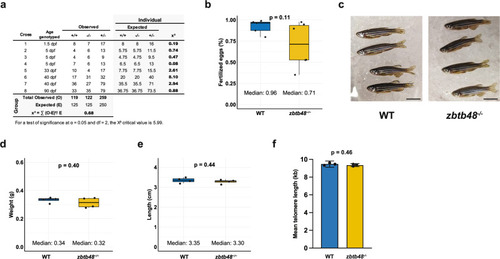
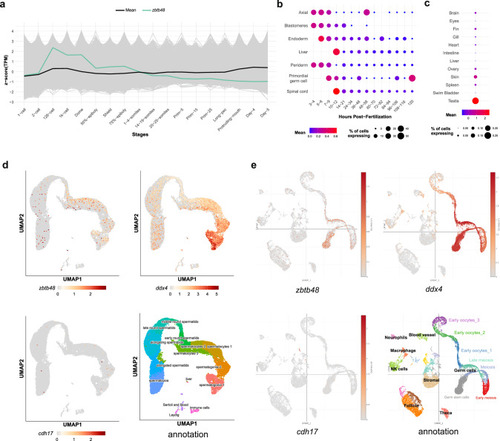
Spatiotemporal expression of |
|
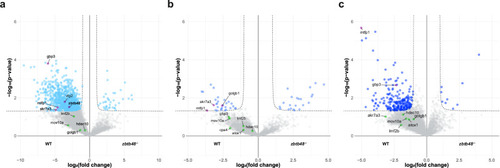
Proteomic analysis of Volcano plots of the proteome analysis of ovaries from 40 dpf ( |