|
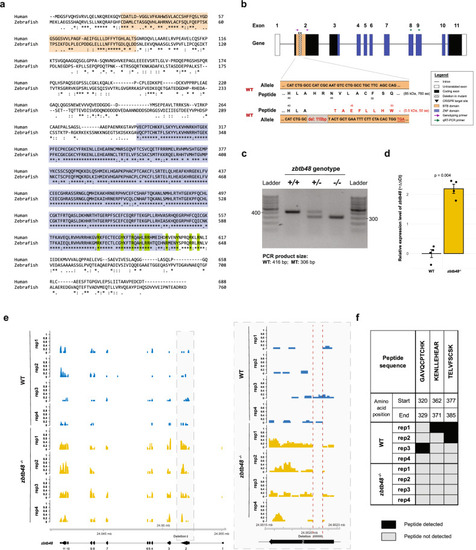
Generation and validation of the zbtb48 CRISPR‒Cas knockout zebrafish line. a Protein sequence alignment of human ZBTB48 and zebrafish Zbtb48 showing greater homology in the telomere-binding zinc finger domain (80% identity; blue) than in the BTB domain (42% identity; beige). The ten amino acids highlighted in green that are involved in telomere binding activity in humans (Zhao et al.) are conserved in zebrafish. b The zebrafish zbtb48 gene consists of 11 exons (not drawn to scale). To create the zbtb48 CRISPR‒Cas knockout zebrafish line, a pair of guide RNAs was designed to target the second exon, resulting in a 110-base pair frameshift deletion with a premature stop codon. c Results of the genotyping of zbtb48 CRISPR‒Cas knockout zebrafish using agarose gel electrophoresis. d Comparison of zbtb48 mRNA levels between zbtb48−/− mutants and their wild-type counterparts via qRT‒PCR of 5 dpf larvae. The experiment was conducted in biological quadruplicate (25 pooled larvae), each with technical triplicates. Error bars represent the standard error of the mean (SEM), and p-values were calculated using two-tailed Welch’s t test. e mRNA sequencing tracks of the zbtb48 gene region in 5 dpf larvae of wild-type and zbtb48−/− mutant larvae. A magnified image of the boxed area is shown on the right. The CRISPR deletion is marked below the track. f Table showing the Zbtb48 peptides detected via mass spectrometry analysis of a telomere pulldown with lysates extracted from 5 dpf wild-type and zbtb48−/− larvae.
|