|
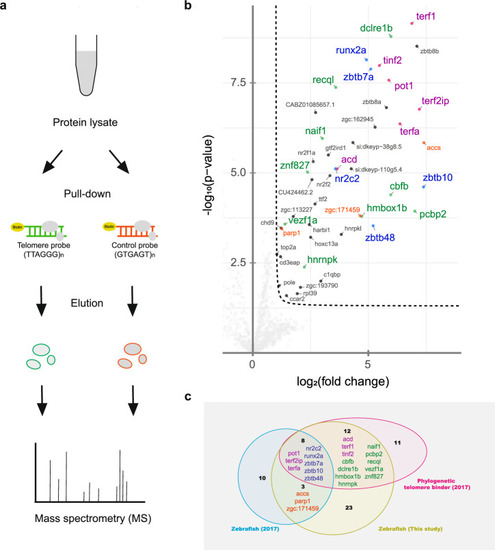
Telomere pull-down with the zebrafish cell line. a Schematic diagram showing an overview of the telomere pull-down experiment using nuclear lysate from the zebrafish fin fibroblast BRF41 cell line. b Volcano plot visualizing the results obtained by label-free quantification of the bound proteins. The pull-down experiment was conducted in quadruplicate using either concatenated telomeric TTAGGG or a control scrambled GTGAGT oligonucleotide as bait. The results were log-transformed and plotted on the x-axis as log2(fold change) and on the y-axis as -log10(p-value). The protein enrichment threshold was set at a fold change >2 and a p-value < 0.05 (Welch’s t test) with c = 0.05. Enriched proteins were annotated with their gene names and color-coded: shelterin subunits (magenta), proteins that were previously found in the study by Kappei et al. to be enriched in zebrafish (orange), and telomere-binding proteins previously identified as phylogenetically conserved in other species (green), which were also enriched in zebrafish (blue). c Comparison of the previous telomere zebrafish pull-down, the phylogenetic conserved binders and the candidates from the current study.
|