- Title
-
Molecularly engineered supramolecular fluorescent chemodosimeter for measuring epinephrine dynamics
- Authors
- Zhao, Y., Mei, Y., Liu, Z., Sun, J., Tian, Y.
- Source
- Full text @ Nat. Commun.
|
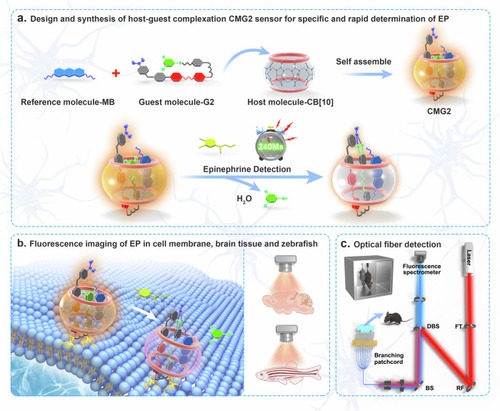
The host-guest GMG2 chemodosimeter for visualizing and quantifying of EP in vitro and in vivo. |
|
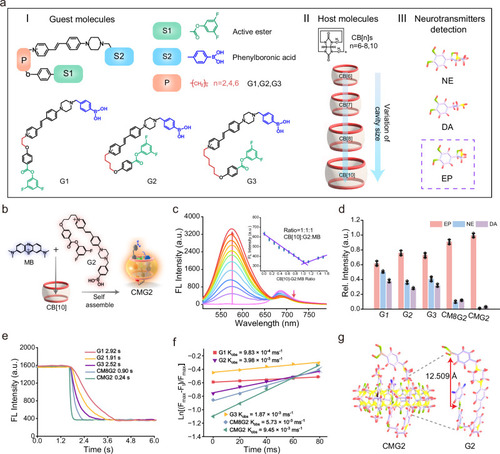
Design and synthesis of host–guest supramolecular fluorescent chemodosimeters for selective and rapid response toward EP. |
|
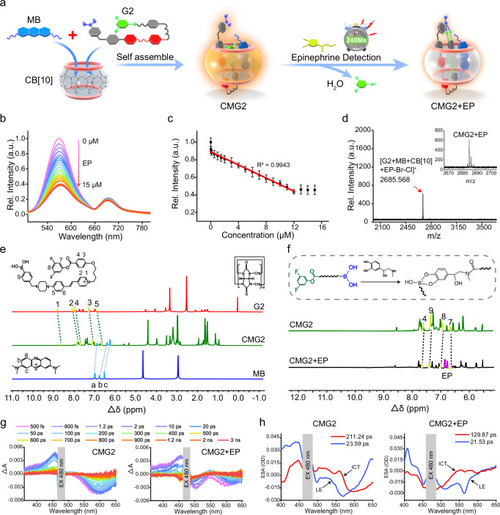
Fluorescence titration of CMG2 toward EP and mechanism evaluation. |
|
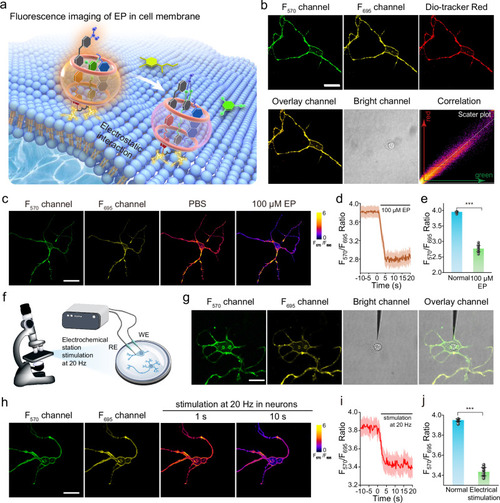
Fluorescence imaging and real-time quantification of EP in neurons. |
|
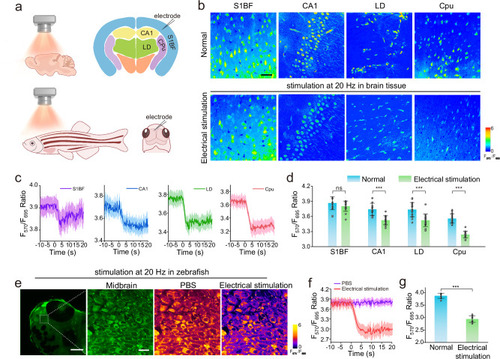
Fluorescence imaging and real-time quantification of EP in brain tissues and zebrafish. |
|
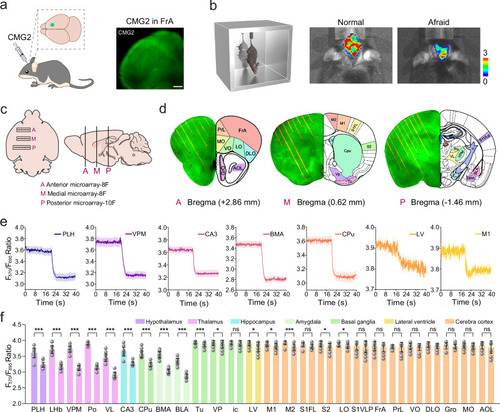
Real-time monitoring and quantifying of EP in 26 brain regions. |