- Title
-
A zebrafish-based in vivo model of Zika virus infection unveils alterations of the glutamatergic neuronal development and NS4A as a key viral determinant of neuropathogenesis
- Authors
- Sow, A.A., Jamadagni, P., Scaturro, P., Patten, S.A., Chatel-Chaix, L.
- Source
- Full text @ PLoS Pathog.
|
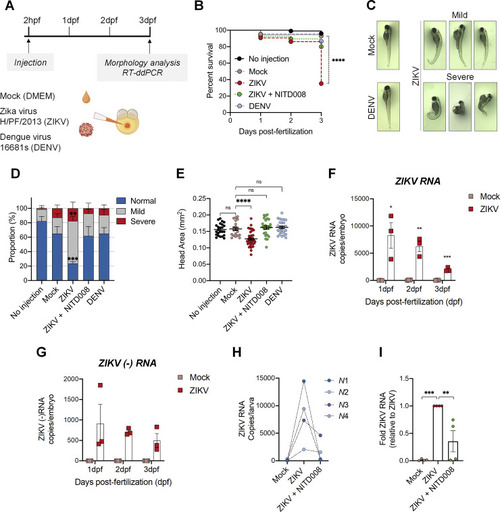
Zika virus replicates efficiently in the zebrafish model and induces morphological defects. (A) Schematic experimental design. Cell medium (mock), DENV viral particles (serotype 2, strain 16681s) or ZIKV viral particles (strain H/PF/2013) were microinjected in the zebrafish yolk at 2 hours post-fertilization (hpf). This schematic was created with |
|
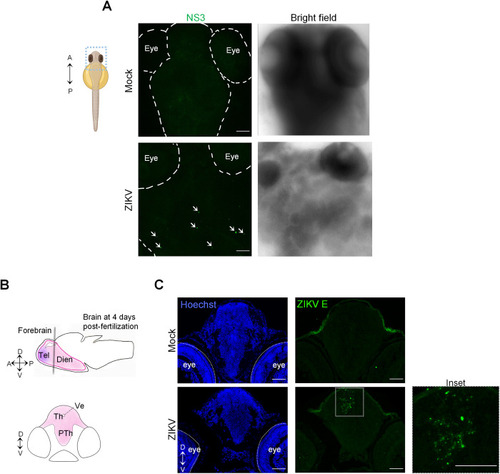
ZIKV injection results in viral protein accumulation in larval brain. (A) At 3 dpf, mock- and ZIKV-infected larvae were fixed and subjected to whole mount immunostaining with anti-NS3 antibodies. Representative pictures of 4 imaged samples are shown. The arrows indicate specific ZIKV NS3 signals. The schematic representation of the zebrafish larva illustrating the region of interest was created with |
|
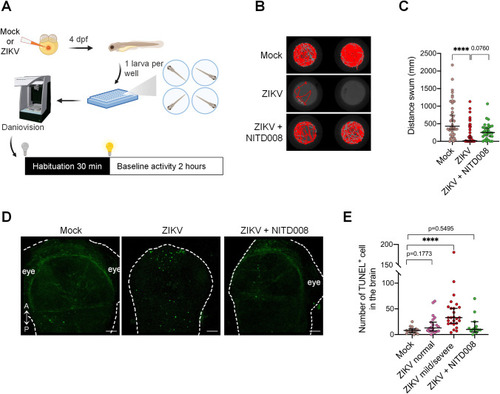
Locomotor defects and brain cell death in developing zebrafish following ZIKV infection. (A) Schematic experimental setup and behavioral analysis (created with |
|
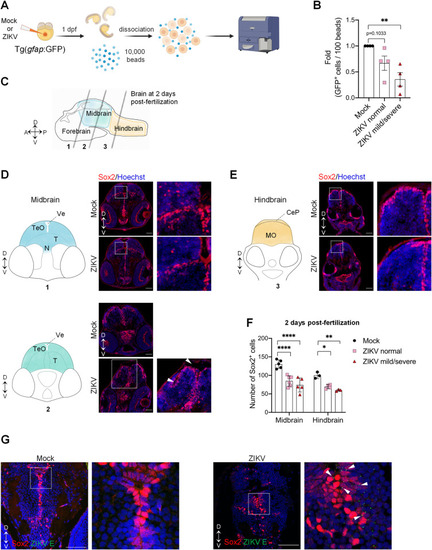
Zika virus targets neural progenitor cells and induces neuropathogenesis in zebrafish larvae. (A) Schematic experimental design (created with |
|
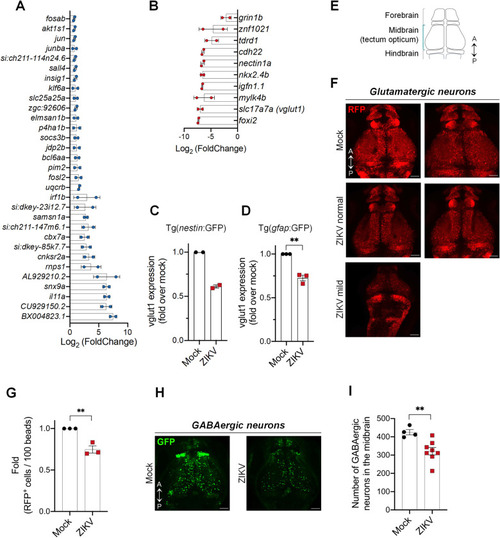
ZIKV infection dysregulates the NPC transcriptome and disrupts the glutamatergic and GABAergic neuronal networks. (A-B) Tg( |
|
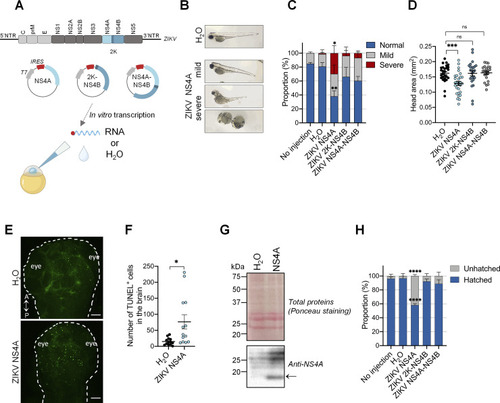
ZIKV NS4A is a major viral determinant in ZIKV pathogenesis (A) Schematic experimental design (created with |