- Title
-
Quantum Dots-caused Retinal Degeneration in Zebrafish Regulated by Ferroptosis and Mitophagy in Retinal Pigment Epithelial Cells through Inhibiting Spliceosome
- Authors
- Zheng, N., Liao, T., Zhang, C., Zhang, Z., Yan, S., Xi, X., Ruan, F., Yang, C., Zhao, Q., Deng, W., Huang, J., Huang, Z.T., Chen, Z.F., Wang, X., Qu, Q., Zuo, Z., He, C.
- Source
- Full text @ Adv Sci (Weinh)
|
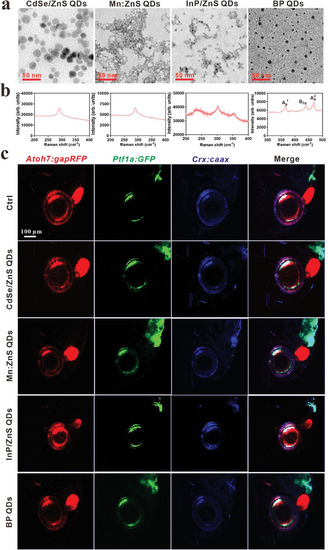
Characterizations of QDs and screening their impacts on the eye development of zebrafish. a) Representative images of QDs by TEM, scale bar, 50 nm. b) The Raman intensity of QDs. c) The lateral views of the effect of QDs (1 mg L−1) on retinal development in Tg ( |
|
InP/ZnS QDs embryonic exposure caused retinal degeneration in zebrafish. a) Schematic illustration of the experimental design. b) Representative images of the distribution of InP/ZnS QDs on zebrafish eye using fluorescence microscopy. Scale bar, 200 µm. c) Determination of indium element content in the retina of zebrafish by ICP‐MS, n = 6, ND: No Detected. d) Representative images of zebrafish from 72 to 168 hpf after exposure. Scale bar, 1 mm. e) Schematic diagram of horizontal (X) and vertical (Y) axes of zebrafish eye. f) The lengths of X and Y axes in the zebrafish eye at 72–168 hpf, n = 20. Representative images of zebrafish retinal histology at 72 hpf g) and 168 hpf h) as assessed by HE staining. Scale bar, 50 µm. i‐j) Representative images of the retinas of Sofa zebrafish at 72 hpf i) and 168 hpf j) using a light‐sheet microscope. The retinal ganglion cell layer (GCL) is labeled by RPF, the inner nuclear layer (INL, composed of AC and HC) is labeled by GFP and CFP, BC is labeled by CFP, and the outer nuclear layer (ONL, composed of PR) labeled by RFP and CFP. k) Scheme of zebrafish eye. l) The representative images of zebrafish eye by optical coherence tomography (OCT) at 21 dpf. The data are presented as mean ± SE. One‐way ANOVA followed by |
|
InP/ZnS QDs embryonic exposure induced many types of cell injury and impaired phototransduction in zebrafish retinas using bulk RNA‐seq. After exposure to InP/ZnS QDs, the zebrafish eyes at 72 hpf were collected to perform RNA‐seq, n = 3. a) The PCA result of control, 0.1 and 1 mg L−1 InP/ZnS QDs. b) Venn diagram of the overlapped DEGs in the 0.1 and 1 mg L−1 groups compared to the control with 0.5‐fold cutoff and |
|
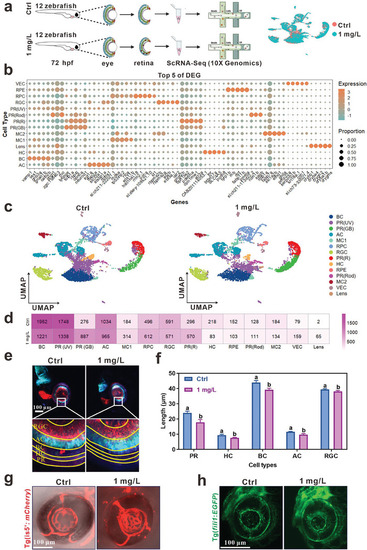
ScRNA‐seq analysis displayed the retinal degenerative damage of zebrafish exposed to InP/ZnS QDs at single cell level. a) Schematic illustration of the scRNA‐seq experimental design. We dissociated the retinas from zebrafish exposed to InP/ZnS QDs from 0.5 to 72 hpf (24 retinas of 12 larvae from each group). b) The top five DEGs of each cluster. c) UMAP plots show the clustering of qualified zebrafish eyes at 72 hpf. Distinct cell types are marked by different colors. d) The cell number of different clusters between control and InP/ZnS QDs‐exposed group. e) The representative images of the retina of Tg ( |
|
The number and function alteration of PR cells in zebrafish after exposure to InP/ZnS QDs. a) Pseudotime trajectory analysis of retinal cell development with Monocle. Each dot is a single cell and its color is agreement with its cluster assignment. b–e) The representative images of PR(Rod), PR(UV), PR(GB), and PR(R) of zebrafish at 72 hpf stained by RHO b), OPN1SW1 c), OPN1MW1 d), and OPN1LW2 e), respectively. f–i) The scheme and gain of optokinetic response (OKR) of 168‐hpf zebrafish in Black‐White f), Blue‐White g), Green‐White h), and Red‐White stripes i), n ≥ 5. The data are presented as mean ± SE. Significant change among different groups ( |
|
InP/ZnS QDs embryonic exposure promoted the differentiation of PR(UV) to PR(RGB) in zebrafish. a‐e) The bioinformatics analysis was based on the data from scRNA‐seq. a) KEGG pathway analysis of the DEGs in PR(UV) cells. b) The GSEA of PR(UV) cells. c) KEGG pathway analysis of the DEGs in PR(RGB) cells. d) The GSEA of PR(RGB) cells. e) The single‐cell RNA velocity of four types of PR cells in the control and InP/ZnS QDs group, respectively, using by cell velocity. The red cycle indicates the differentiation of PR(UV) to PR(RGB). f) Fluorescence triple staining assay (TSA) was used to track PR(UV), PR(R), and PR(GB), respectively. The white arrowheads display the co‐location of PR(UV) with PR(R) or PR(GB), suggesting the differentiation of PR(UV) to PR(RGB) caused by InP/ZnS QDs exposure. The images were captured by confocal microscopy. Scale bar, 10 µm. |
|
InP/ZnS QDs, endocytosed by RPE cells, caused retinal degenerative damage involved in mitophagy and ferroptosis due to spliceosome inhibition. a) The scheme shows the interaction between RPE and PR.[ |
|
Mechanism of retinal degeneration induced by InP/ZnS QDs in zebrafish through inhibiting |