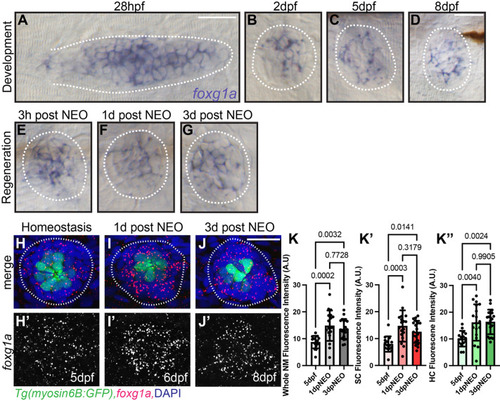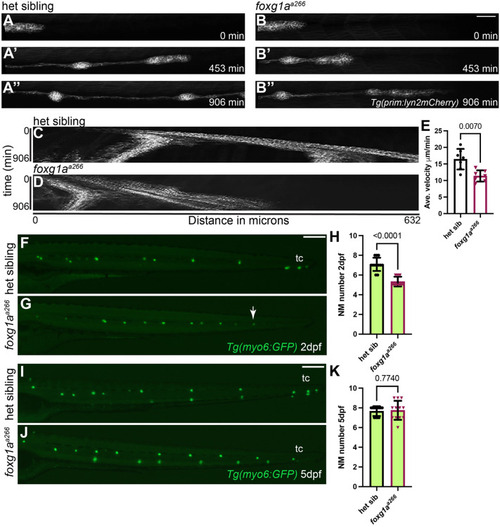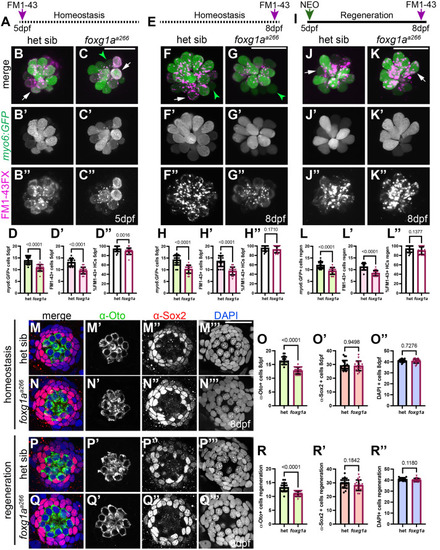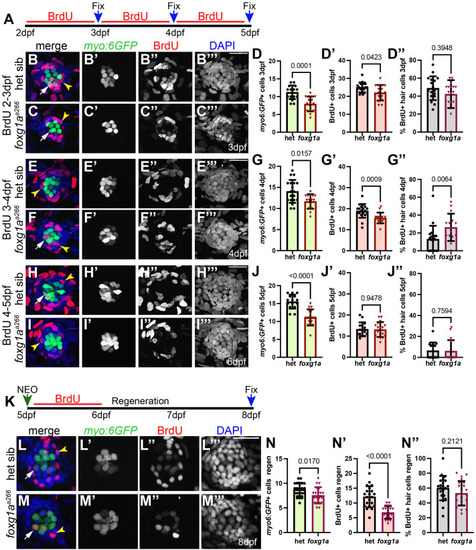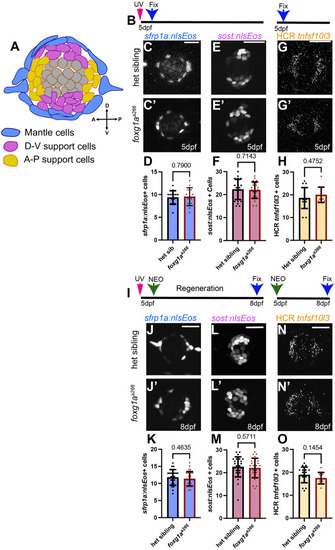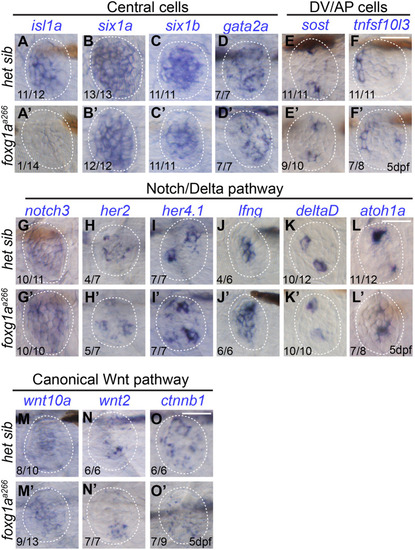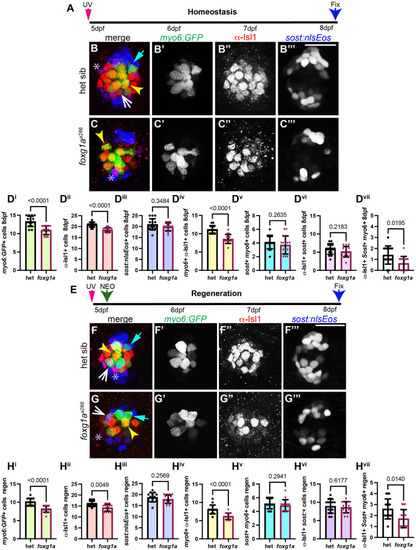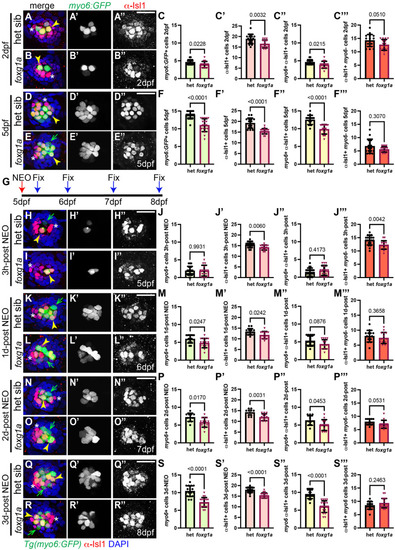
α-Islet1 antibody-labeled cells are reduced in foxg1aa266 mutants during development and regeneration. Confocal projections of neuromasts showing hair cells labeled with myo6:GFP (green), central cells and hair cells labeled with α-Isl1 antibody (red), and nuclei labeled with DAPI (blue). Examples of hair cells that are both myo6:GFP+ and α-Isl1+ are marked with yellow arrowheads, hair cells that are myo6:GFP+ and α-Isl1- are marked by green arrows, and cells that are α-Isl1+ and myo6:GFP- are marked with white asterisks. (A-B″) Heterozygous sibling and foxg1aa266 mutants neuromasts at 2 dpf. (C-C′″) Quantification of myo6:GFP+ hair cells, α-Isl1+ cells, cells labeled with both myo6:GFP and α-Isl1, and cells labeled with only α-Isl1, n=17 NMs (nine embryos) heterozygous siblings and 15 NMs (eight embryos) foxg1aa266 mutants. (D-E″) Heterozygous sibling and foxg1aa266 mutant neuromasts at 5 dpf. (F-F′″) Quantification of myo6:GFP+ hair cells, α-Isl1+ cells, cells labeled with both myo6:GFP and α-Isl1, and cells labeled with only α-Isl1, n=16 neuromasts (eight larvae) heterozygous siblings and 18 neuromasts (nine larvae) foxg1aa266 mutants. (G) Timeline of NEO exposure at 5 dpf and fixation at 3 h-post NEO (5 dpf), 1 day-post NEO (6 dpf), 2 days-post NEO (7 dpf), and complete regeneration (8 dpf). (H-I″) Heterozygous sibling and foxg1aa266 mutants neuromasts at 3 h-post NEO (5 dpf). (J-J′″) Quantification of myo6:GFP+ hair cells, α-Isl1+ cells, cells labeled with both myo6:GFP and α-Isl1, and cells labeled with only α-Isl1, n=20 neuromasts (ten larvae) heterozygous siblings and 15 neuromasts (eight larvae) foxg1aa266 mutants. (K-L″) Heterozygous sibling and foxg1aa266 mutant neuromasts at 1 day-post NEO (6 dpf). (M-M′″) Quantification of myo6:GFP+ hair cells, α-Isl1+ cells, cells labeled with both myo6:GFP and α-Isl1, and cells labeled with only α-Isl1, n=14 neuromasts (seven larvae) heterozygous siblings and 15 neuromasts (eight larvae) foxg1aa266 mutants. (N-O″) Heterozygous sibling and foxg1aa266 mutants neuromasts at 2 days-post NEO (7 dpf). (P-P′″) Quantification of myo6:GFP+ hair cells, α-Isl1+ cells, cells labeled with both myo6:GFP and α-Isl1, and cells labeled with only α-Isl1, n=11 neuromasts (seven larvae) heterozygous siblings and 13 neuromasts (eight larvae) foxg1aa266 mutants. (Q-R″) Heterozygous sibling and foxg1aa266 mutants neuromasts at complete regeneration NEO (8 dpf). (S-S′″) Quantification of myo6:GFP+ hair cells, α-Isl1+ cells, cells labeled with both myo6:GFP and α-Isl1, and cells labeled with only α-Isl1, n=17 neuromasts (nine larvae) heterozygous siblings and 19 neuromasts (nine larvae) foxg1aa266 mutants. All data presented at mean±s.d., Mann–Whitney U-test. Scale bars: 20 µm.
|