- Title
-
Lysosomal membrane protein TMEM106B modulates hematopoietic stem and progenitor cell proliferation and differentiation by regulating LAMP2A stability
- Authors
- Guo, D., Xiong, H., Yang, Z., Zhang, R., Shi, P., Yao, Y., Liu, M., Xu, C., Wang, Q.K.
- Source
- Full text @ FASEB J.
|
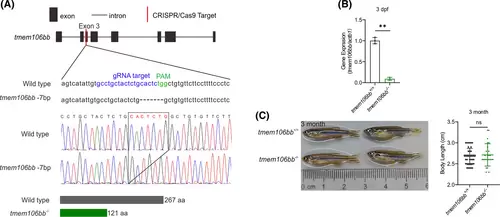
Generation and characterization of tmem106bb KO zebrafish using CRISPR/Cas9 genome editing. (A) Strategy of CRISPR/Cas9 editing of tmem106bb with a site-specific targeting sgRNA for CRISPR/Cas9 cleavage in exon 3. Genomic DNA sequence analysis identified a mutant zebrafish line with a 7-bp deletion on exon 3, a frameshift mutation that results in a truncated Tmem106b protein with N-terminal 121 amino acids. (B) Near absence of tmem106bb mRNA in 3-dpf tmem106bb−/− KO embryos compared with wild-type tmem106bb+/+ embryos as revealed by RT-qPCR analysis (n = 3 samples/group with 20 embryos in each sample). (C) Comparison of morphology and length of 3-month adult tmem106bb−/− and compared with wild-type tmem106bb+/+ (n = 25–38 per group). **p < .01, ns, no significance; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and paired (B) or unpaired (C) Student's t tests with two tails. |
|
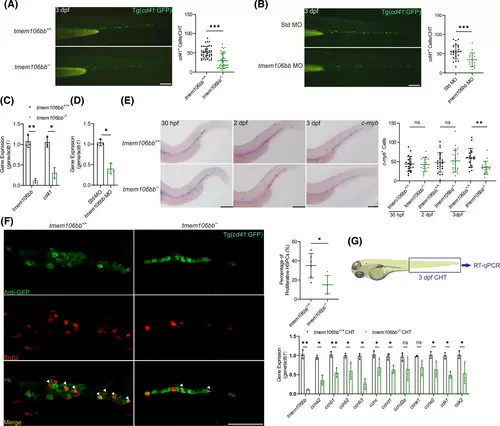
Both KO and knockdown of tmem106bb reduce HSPC proliferation. (A) Signal of cd41+ HSPCs was significantly reduced in CHT of 3 dpf tmem106bb−/−/Tg(cd41:GFP) embryos compared with control tmem106bb+/+/Tg(cd41:GFP) embryos (n = 37–39 embryos per group; scale bar: 200 μm). (B) Signal of cd41+ HSPCs was significantly reduced in CHT of 3 dpf Tg(cd41:GFP) embryos injected with tmem106bb-MO compared with those injected with control Std-MO (n = 16–26 embryos per group; scale bar: 200 μm). (C) RT-qPCR analysis showed that cd41 expression was significantly decreased in tmem106bb−/− embryos compared with wild-type tmem106bb+/+ embryos at 3 dpf (n = 3 samples per group with 50 embryos in each sample). (D) RT-qPCR analysis showed that cd41 expression was significantly decreased in 3 dpf embryos injected with tmem106bb-MO compared with those injected with control Std-MO (n = 3 samples per group with 50 embryos in each sample). (E) WISH for HSPC marker c-myb with tmem106bb−/− and tmem106bb+/+ embryos at 30 hpf, 2 and 3 dpf (n = 16–24 embryos per group; scalebar: 200 μm). (F) Cell proliferation analysis with BrdU staining of 3 dpf tmem106bb−/−/Tg(cd41:GFP) and control tmem106bb+/+/Tg(cd41:GFP) embryos. The number of proliferating HSPCs (white triangle arrows) was significantly reduced by tmem106bb KO (n = 6 embryos per group; scale bar: 50 μm). Green signals represent cd41+ HSPCs, red signals indicate BrdU-labeled proliferating cells and yellow signals represent proliferating HSPCs. (G) RT-qPCR analysis for genes involved in cell cycle regulation with CHT samples of 3 dpf tmem106bb−/− and control tmem106bb+/+ embryos (n = 3 samples per group with 50 embryos in each sample). *p < .05, **p < .01, ***p < .001, ns, no significance; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and unpaired (A, B, E, F) or paired (C, D, G) student's t tests with two tail. |
|
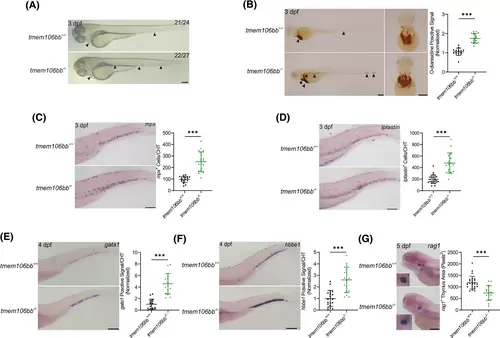
KO of tmem106bb impairs definitive hematopoiesis. (A) Appearance of embryos showed increased red blood cells in tmem106bb−/− embryos at 3 dpf (n = 24–27 embryos per group; scale bar: 200 μm). The black arrowheads indicate red blood cells. (B) O-dianisidine staining showed a significantly increased signal for red blood cells in tmem106bb−/− embryos at 3 dpf (n = 15–18 embryos per group; scale bar: 200 μm). The black arrowheads indicate red blood cells. (C) WISH for mpx showed that myeloid differentiation was significantly increased in tmem106bb−/− embryos at 3 dpf (n = 19–20 embryos per group; scale bar: 200 μm). (D) WISH for lplastin showed that myeloid differentiation was significantly increased in tmem106bb−/− embryos at 3 dpf (n = 17–26 embryos per group; scale bar: 200 μm). (E) WISH for gata1 showed that erythroid differentiation was significantly increased in tmem106bb−/− embryos at 4 dpf (n = 16-embryos per group; scale bar: 200 μm). (F) WISH for hbbe1 showed that erythroid differentiation was significantly increased in tmem106bb−/− embryos at 4 dpf (n = 16–20 embryos per group; scale bar: 200 μm). (G) WISH for rag1 showed that lymphoid differentiation was significantly reduced in tmem106bb−/− embryos at 5 dpf (n = 16–26 embryos per group; scale bar: 200 μm). ***p < .001; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and unpaired student's t tests with two tails. |
|
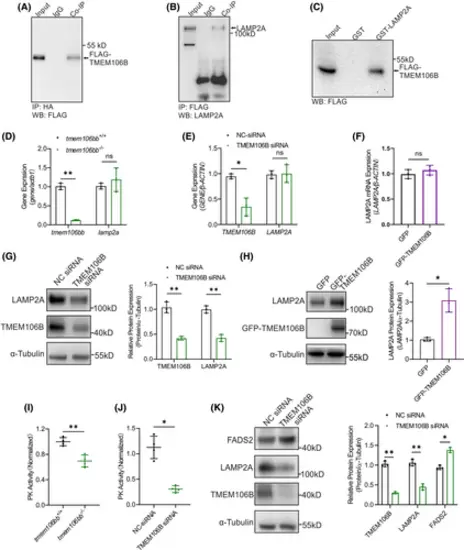
TMEM106B interacts with LAMP2A and regulates LAMP2A protein level. (A, B) Co-IP showing the interaction between TMEM106B and LAMP2A in HeLa cells with co-expression of FLAG-TMEM106B and HA-LAMP2A. (C) GST pull-down assays showing the interaction between GST-LAMP2A and FLAG-tagged wild-type TMEM106B. (D–F) RT-qPCR analysis showed that the mRNA level of lamp2a was not affected by tmem106bb KO in tmem106bb−/− embryos 3 dpf (n = 3 samples per group with 20 embryos in each sample), knockdown with TMEM106B siRNA or overexpression of TMEM106B in HeLa cells (n = 3 samples per group). (G, H) Western blot analysis showed that the LAMP2A protein level was significantly decreased by knockdown of TMEM106B with siRNA compared with NC siRNA in HeLa cells or increased with overexpression of TMEM106B in HeLa cells (n = 3 samples per group). (I) Pyruvate kinase activity was significantly decreased in tmem106bb−/− embryos at 3 dpf (n = 4 samples per group with 100 embryos in each sample). (J) Pyruvate kinase activity was significantly decreased in HeLa cells with knockdown of TMEM106B by siRNA compared with cells transfected with NC siRNA (n = 4 samples per group). (K) Western blot analysis showed that the FASD2 protein level was significantly increased in HeLa cells with knockdown of TMEM106B by siRNA compared with cells transfected with NC siRNA (n = 3 samples per group). *p < .05, **p < .01, ns, no significance; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and paired student's t tests with two tails. |
|
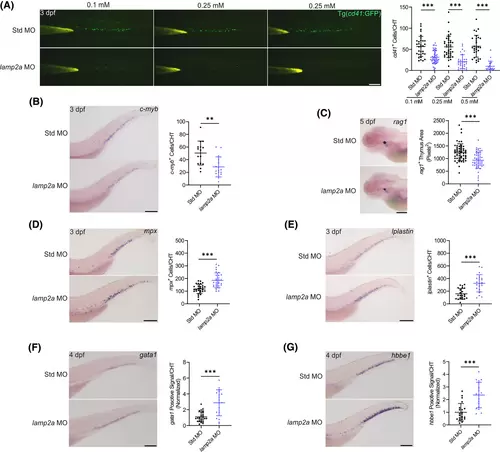
Knockdown of lamp2a impairs the proliferation and differentiation capability of HSPCs. (A) Signal of cd41+ HSPCs was significantly reduced in CHT of 3 dpf Tg(cd41:GFP) embryos injected with lamp2a-MO compared with those with control Std-MO in a dose-dependent manner (n = 19–48 embryos per group; scale bar: 200 μm). (B) WISH for c-myb showed that the HSPC signal was significantly reduced in 3 dpf embryos injected with lamp2a-MO compared with Std-MO (n = 28–39 embryos per group; scale bar: 200 μm). (C) WISH for rag1 showed that lymphoid differentiation was significantly reduced in 5 dpf embryos injected with lamp2a-MO compared with Std-MO (n = 51–56 embryos per group; scale bar: 200 μm). (D) WISH for mpx showed that myeloid differentiation was significantly increased in 3 dpf embryos injected with lamp2a-MO compared with Std-MO (n = 29–30 embryos per group; scale bar: 200 μm). (E) WISH for lplastin showed that myeloid differentiation was significantly increased in 3 dpf embryos injected with lamp2a-MO compared with Std-MO (n = 21–22 embryos per group; scale bar: 200 μm). (F) WISH for gata1 showed that erythroid differentiation was significantly increased in 4 dpf embryos injected with lamp2a-MO compared with Std-MO (n = 18–26 embryos per group; scale bar: 200 μm). (G) WISH for hbbe1 showed that erythroid differentiation was significantly increased in 4 dpf embryos injected with lamp2a-MO compared with Std-MO (n = 18–22 embryos per group; scale bar: 200 μm). **p < .01, ***p < .001; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and unpaired student's t tests with two tail. |
|
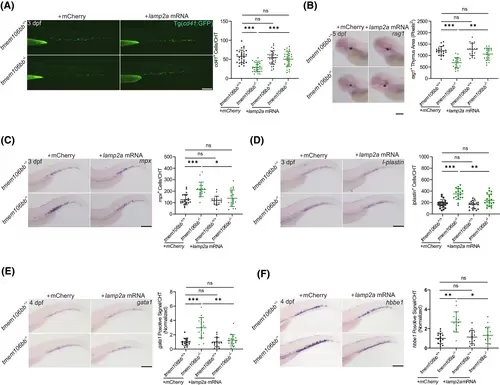
Overexpression of lamp2a rescues the impaired definitive hematopoiesis in tmem106bb KO embryos. (A) Injection of lamp2a mRNA (mCherry RNA as control) rescued the reduced signal of cd41+ HSPCs in CHT of 3 dpf tmem106bb−/−/Tg(cd41:GFP) embryos (n = 29 embryos per group; scale bar: 200 μm). (B) WISH for rag1 showed that injection of lamp2a mRNA (mCherry RNA as control) rescued the decreased lymphoid differentiation in tmem106bb morphants (n = 19–20 embryos per group; scale bar: 200 μm). (C) WISH for mpx showed that injection of lamp2a mRNA (mCherry RNA as control) reversed the increased myeloid differentiation in tmem106bb morphants (n = 17–22 embryos per group; scale bar: 200 μm). (D) WISH for lplastin showed that injection of lamp2a mRNA (mCherry RNA as control) reversed the increased myeloid differentiation in tmem106bb morphants (n = 18–21 embryos per group; scale bar: 200 μm). (E) WISH for gata1 showed that injection of lamp2a mRNA (mCherry RNA as control) reversed the increased erythroid differentiation in tmem106bb morphants (n = 16-embryos per group; scale bar: 200 μm). (F) WISH for hbbe1 showed that injection of lamp2a mRNA (mCherry RNA as control) reversed the increased erythroid differentiation in tmem106bb morphants (n = 16-embryos per group; scale bar: 200 μm). *p < .05, **p < .01, ***p < .001, ns, no significance; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and two-way anova followed by a Sidak correction post hoc test. |
|
TMEM106B regulates the degradation of LAMP2A by influencing the interaction between LAMP2A and CTSA. (A) LAMP2A degradation with or without overexpression of TMEM106B in HeLa cells. HeLa cells were transfected with HA-LAMP2A with or without GFP-TMEM106B, treated with 10 μg/mL cycloheximide (CHX) for specified times, and used for Western blot analysis. Overexpression of TMEM106B slowed down the degradation of LAMP2A. (B) Western blot analysis showed that the expression of mature-CTSA was not affected by knockdown of TMEM106B with siRNA compared with NC siRNA in HeLa cells (n = 3 samples per group). (C) Co-IP analysis showing the interaction between CTSA and LAMP2A in HeLa cells with co-expression of FLAG-CTSA and HA-LAMP2A. (D) Co-IP analysis showing the interaction of CTSA and LAMP2A with or without the overexpression of TMEM106B. The graph on the right shows quantification of the binding of CTSA to LAMP2A (n = 4 samples per group). *p < .05, **p < .01, ns, no significance; Data were shown as mean ± SD; Statistical analysis was performed using a Shapiro–Wilk test for samples with normal distribution and paired student's t tests with two tails. |
|
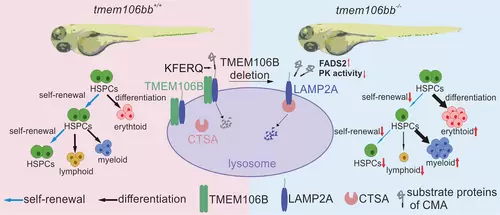
Schematic working model for the role of Tmem106bb in the maintenance of HSPC numbers and functions during zebrafish embryogenesis. Tmem106bb is required for the self-renewal and maintenance of differentiation potentials of HSPCs. In tmem106bb−/− KO zebrafish, HSPCs exhibit impaired self-renewal and disrupted differentiation. The underlying molecular mechanism is that TMEM106B acts as a chaperone for LAMP2A, and loss of TMEM106B increases the degradation of LAMP2A, which impairs CMA and causes abnormal HSPC proliferation and differentiation. |