- Title
-
Oxygen enhances antiviral innate immunity through maintenance of EGLN1-catalyzed proline hydroxylation of IRF3
- Authors
- Liu, X., Tang, J., Wang, Z., Zhu, C., Deng, H., Sun, X., Yu, G., Rong, F., Chen, X., Liao, Q., Jia, S., Liu, W., Zha, H., Fan, S., Cai, X., Gui, J.F., Xiao, W.
- Source
- Full text @ Nat. Commun.
|
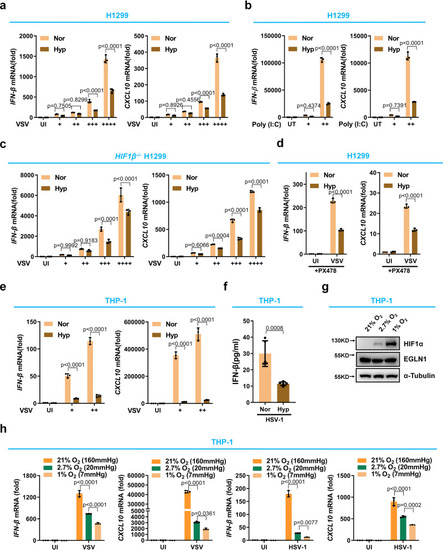
Hypoxia suppresses antiviral gene expression in response to viral infection independently of HIF signaling. |
|
|
|
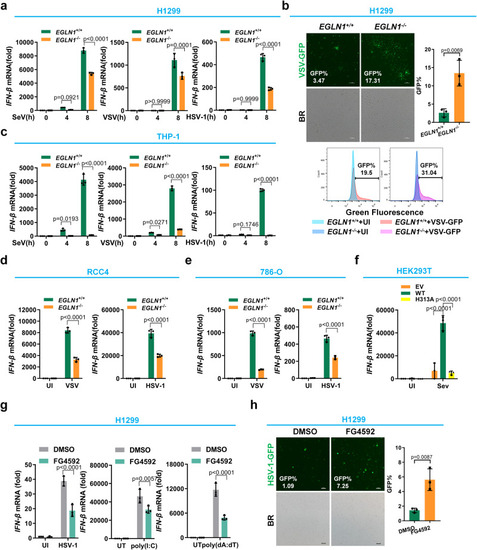
Disruption of |
|
Disruption or inhibition of |
|
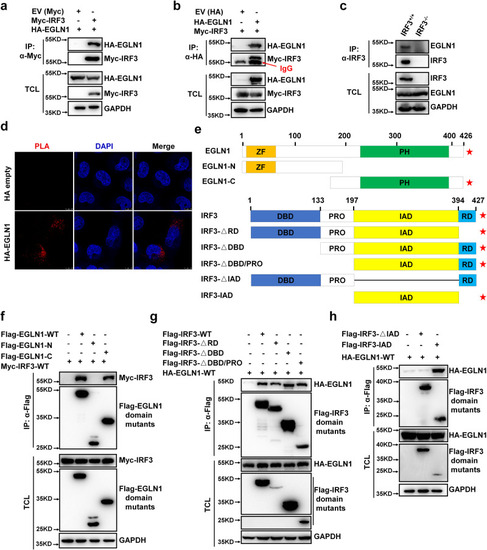
EGLN1 interacts with IRF3. |
|
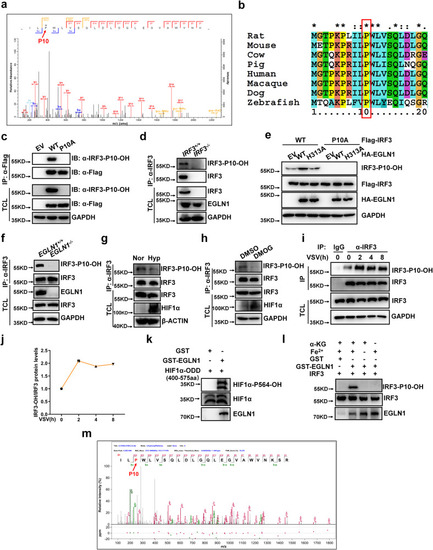
EGLN1 hydroxylates IRF3 at proline 10. |
|
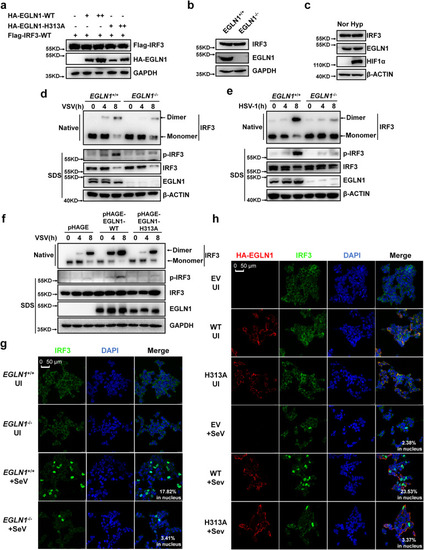
EGLN1 enhances IRF3 phosphorylation, dimerization, and nuclear translocation. |
|
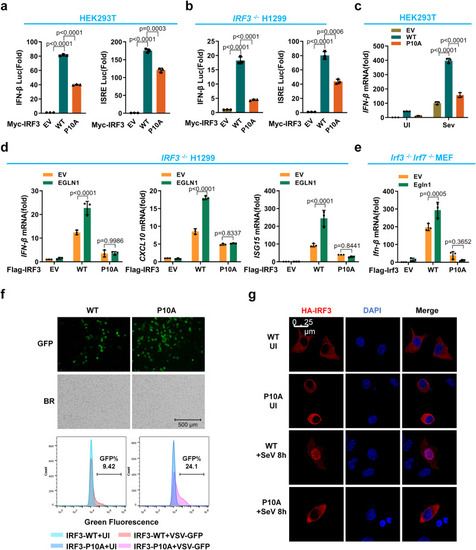
Hydroxylation of IRF3 at proline 10 enhances IRF3 activation and nuclear translocation in cellular antiviral immune responses. |
|
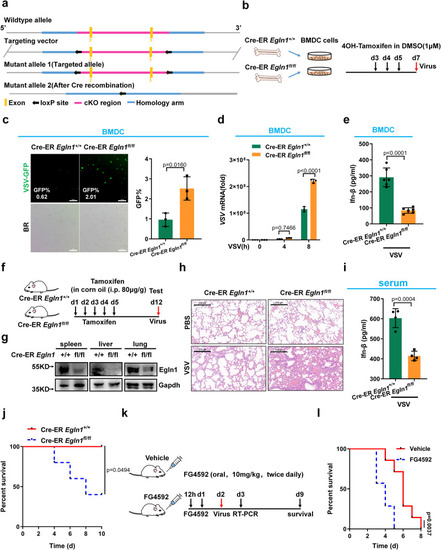
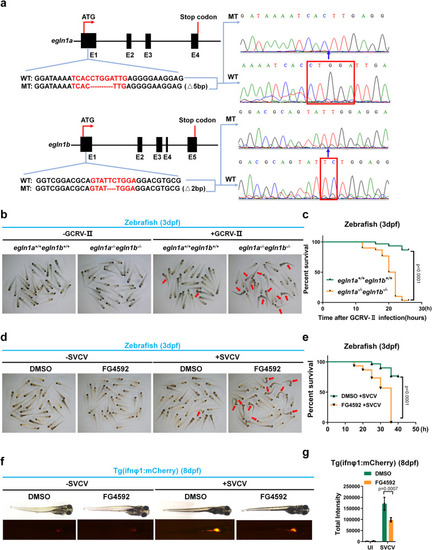
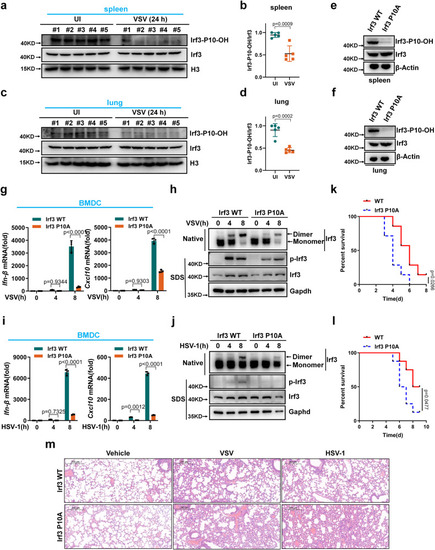
Irf3 prolyl hydroxylation deficiency attenuates antiviral innate immunity in mice. |