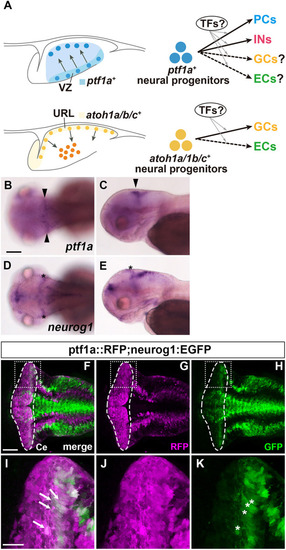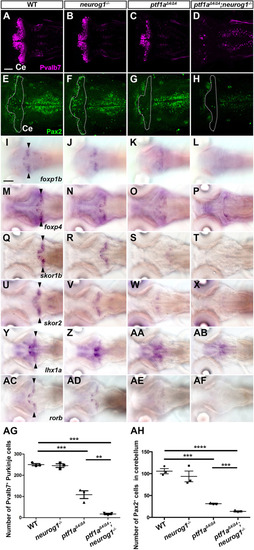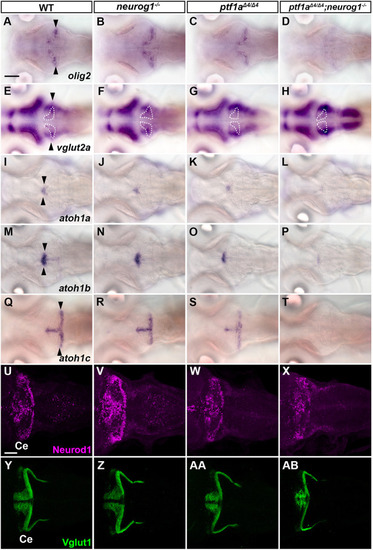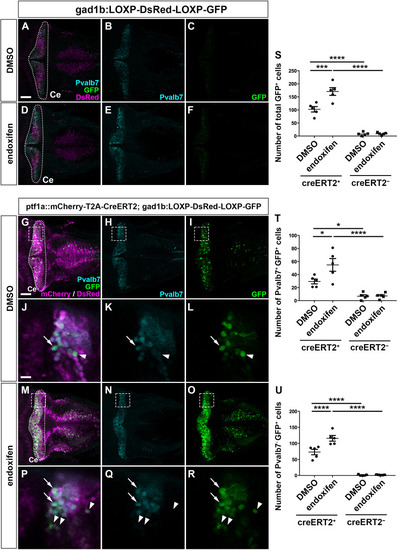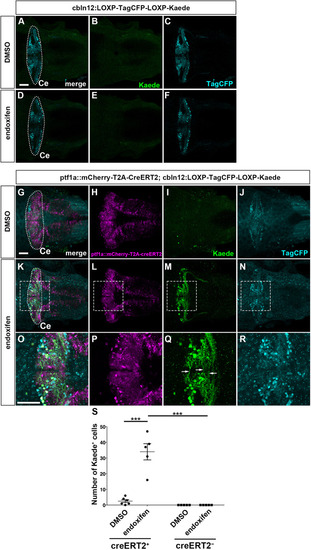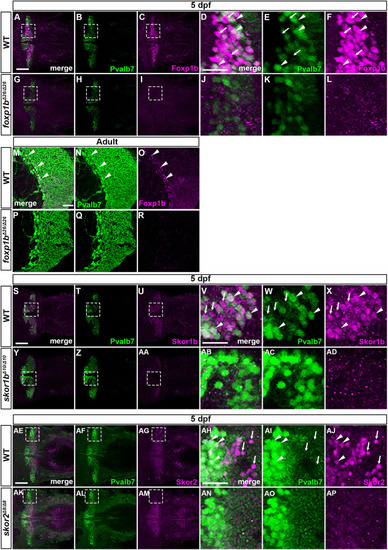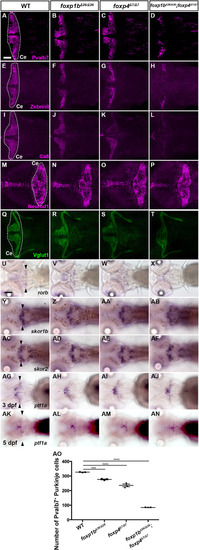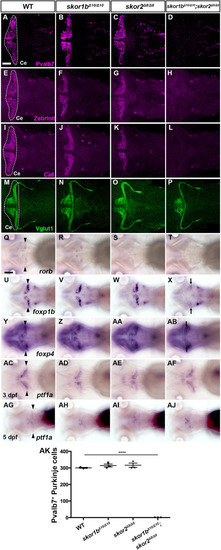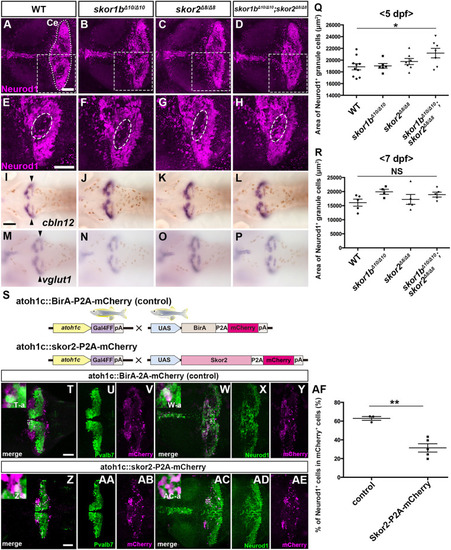
Suppression of granule cell fates by Skor1b/2 and Foxp1b/4. (A-H) Expression of Neurod1 in 5 dpf WT, skor1b, skor2 and skor1b;skor2 mutant larvae, revealed by immunostaining. The cerebellum region is surrounded by a dotted line. (E-H) Higher magnification views of the boxed areas in A-D. Neurod1-expressing GCs were absent in the central areas of the cerebellum (marked by dashed circles) of WT, skor1b and skor2 mutant larvae, but present in the entire cerebellum of skor1b;skor2 mutant larvae. (I-P) Expression of the mature GC marker genes cbln12 and vglut1 in the cerebellum, revealed by in situ hybridization. (Q,R) Area of Neurod1+ GCs in the cerebellum of 5 dpf (Q) or 7 dpf (R) WT, skor1b, skor2 and skor1b;skor2 mutants. *P<0.05 (ANOVA with Tukey's multiple comparison test). (S) Diagram of ectopic expression of biotin ligase A (BirA, control) or Skor2 in GC progenitors. (T-AE) Misexpression of Skor2 in atoh1c-expressing neural progenitors. 5 dpf Tg(atoh1c:Gal4FF);Tg(UAS-hsp70l:BirA-P2A-mCherry) or Tg(atoh1c:Gal4FF);Tg(UAS:HA-skor2-P2A-mCherry) larvae, which express BirA/mCherry or Skor2/mCherry in the GC lineage, were immunostained with anti-RFP/mCherry (magenta), and Pvalb7 (green; T-V,Z-AB) or Neurod1 (green; W-Y,AC-AE) antibodies. Dorsal views with anterior to the left (A-P,T-AE). Insets (T-a,W-a,Z-a,AC-a) show higher magnification views of the boxed areas in T,W,Z,AC. Scale bars: 50 μm (in A, for A-D; in E, for E-H; in T, for T-Y; in Z for Z-AE); 100 μm (in I, for I-P). (AF) Percentage of Neurod1+ cells out of total mCherry+ cells in the cerebellum of larvae expressing BirA (control) or Skor2. **P<0.01 (unpaired two-tailed Student's t-test). Data are shown as mean±s.e.m. with individual values indicated (Q,R,AF).
|