- Title
-
Antagonistic interactions safeguard mitotic propagation of genetic and epigenetic information in zebrafish
- Authors
- Lawir, D.F., Soza-Ried, C., Iwanami, N., Siamishi, I., Bylund, G.O., O Meara, C., Sikora, K., Kanzler, B., Johansson, E., Schorpp, M., Cauchy, P., Boehm, T.
- Source
- Full text @ Commun Biol
|
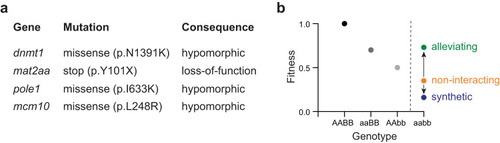
Study design. |
|
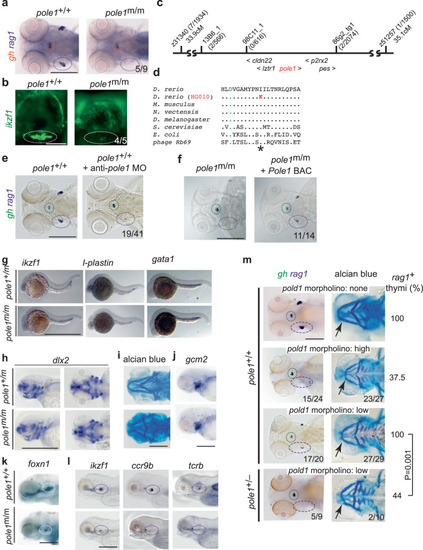
Characterization of a zebrafish |
|
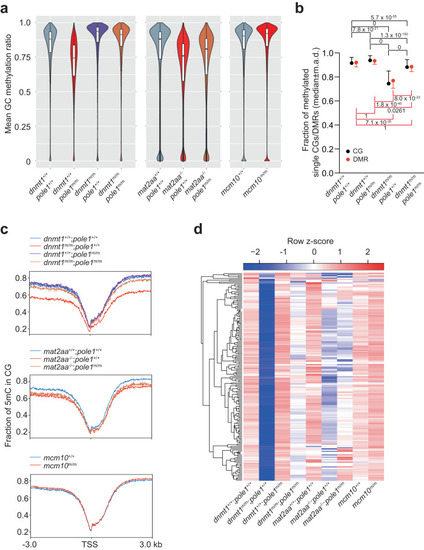
Global changes in DNA methylation patterns in single and double mutant zebrafish. |
|
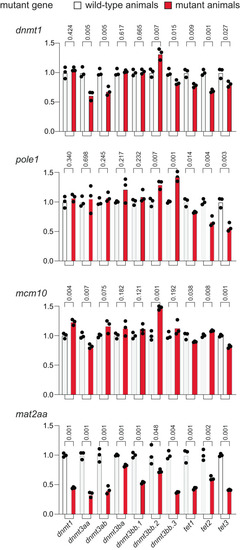
Expression analysis of genes associated with the DNA methylation process. qPCR analysis was performed on 5 dpf embryos of wild-type and mutants ( |
|
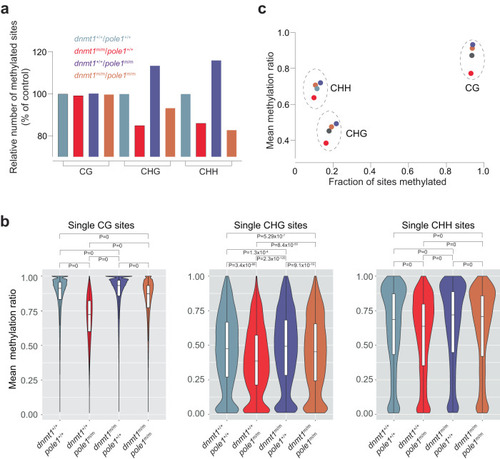
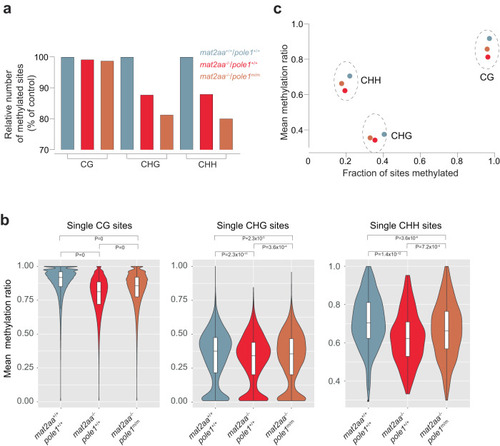
Alterations in non-CG methylation patterns in |
|
Alterations in non-CG methylation patterns in |
|
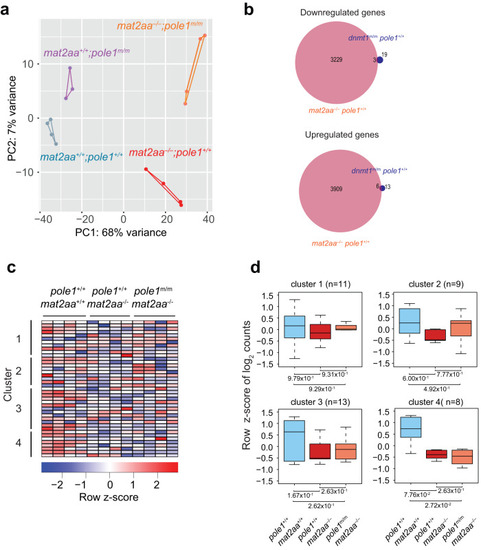
Transcriptional landscapes in |
|
Transcriptional landscapes in |
|
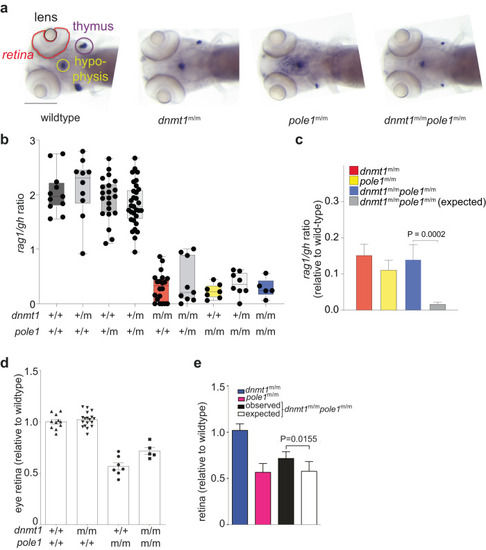
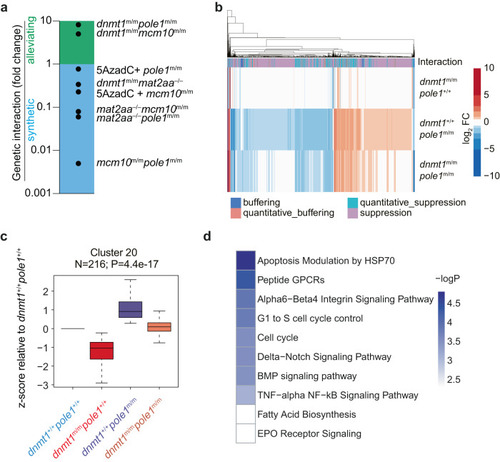
Epistasis analysis of |
|
Genetic interaction analysis. |