- Title
-
PRDM1 DNA-binding zinc finger domain is required for normal limb development and is disrupted in split hand/foot malformation
- Authors
- Truong, B.T., Shull, L.C., Lencer, E., Bend, E.G., Field, M., Blue, E.E., Bamshad, M.J., Skinner, C., Everman, D., Schwartz, C.E., Flanagan-Steet, H., Artinger, K.B.
- Source
- Full text @ Dis. Model. Mech.
|
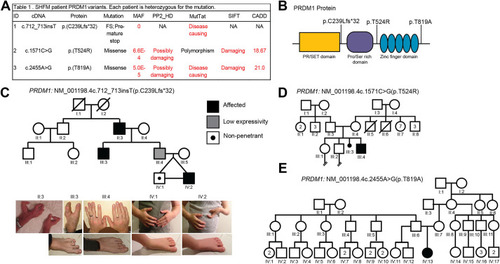
PRDM1 variants of unknown significance identified in families with split hand/foot malformation (SHFM). (A) Table showing PRDM1 variants and predictions of pathogenicity based on various bioinformatics tools. CADD, Combined Annotation-Dependent Depletion; FS, frameshift; MAF, minor allele frequency; MutTat, MutationTaster; NA, not applicable; PP2_HD, Polymorphism Phenotyping v2 HumDiv; SIFT, Sorting Intolerant From Tolerant. (B) Schematic of PRDM1 structure and location of variants identified in individuals with SHFM. (C) Pedigree for family with PRDM1 variant #1, c.712_713insT (p.C239Lfs*32). The symbols representing affected individuals are shaded. Standard pedigree symbols are used. The variant is inherited in an autosomal-dominant manner with incomplete penetrance and variable expressivity. Photographs of the limbs of the individuals in the family are also shown. (D) Pedigree for family with PRDM1 variant #2, c.1571C>G (p.T524R). (E) Pedigree for family with PRDM1 variant #3, c.2455A>G (p.T819A). Variants #2 and #3 are de novo. |
|
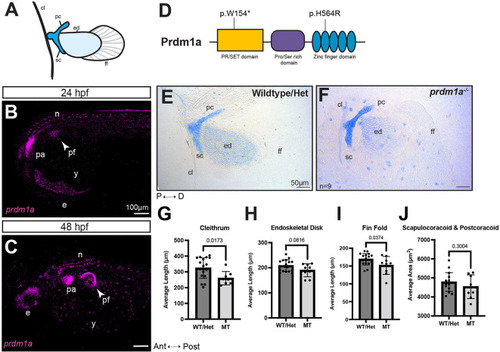
prdm1a−/− zebrafish mutants have hypoplastic pectoral fins. (A) Cartoon of pectoral fin bud at 4 dpf. (B,C) Lateral view of whole-mounted embryos after hybridization chain reaction (HCR) for prdm1a at (B) 24 (n=3) and (C) 48 hpf (n=28). Anterior is to the left. Images are maximum projections of the whole embryo. Arrowheads point to the pectoral fin bud. Scale bars: 100 µm. (D) Schematic of Prdm1a protein. The prdm1am805/m805 allele causes a premature stop codon in the SET domain and is a presumed null mutation (p.W154*). The hypomorphic prdm1atp39/tp39 allele is a missense mutation in the second zinc finger (p.H564R). (E,F) Representative images of Alcian Blue-stained pectoral fins for (E) WT/heterozygous (n=18) and (F) prdm1a−/− mutants (n=9) at 4 dpf. Scale bars: 50 µm. (G-J) The average lengths of the (G) cleithrum, (H) endoskeletal disk and (I) fin fold and (J) the average area of the scapulocoracoid/postcoracoid were measured. Each dot represents one independent biological replicate. Averages were compared with an unpaired, two-tailed independent Student's t-test. prdm1a−/− mutants had a shorter cleithrum (P=0.0173), endoskeletal disk (P=0.0816) and fin fold (P=0.0374). Error bars represent the mean±s.d. The representative images in E and F are also shown in Fig. S1, where the WT and heterozygous fins are analyzed separately. Ant, anterior; cl, cleithrum; D, distal; e, eye; ed, endoskeletal disk; ff, fin fold; Het, heterozygous; hpf, hours post fertilization; MT, prdm1a−/− mutant; n, neurons; P, proximal; pa, pharyngeal arches; pc, postcoracoid; pf, pectoral fin; Post, posterior; sc, scapulocoracoid; WT, wild-type; y, yolk. |
|
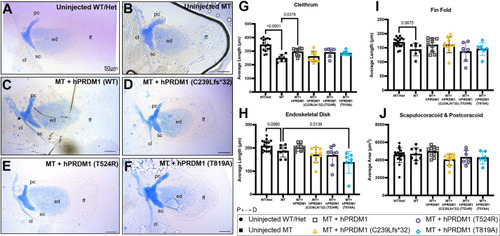
Transient overexpression of SHFM hPRDM1 variants fails to rescue the pectoral fin in prdm1a−/− mutants. prdm1a+/− heterozygous fish were intercrossed and injected with the hPRDM1 WT and SHFM variant mRNAs at the single-cell stage. Injected larvae were collected at 4 dpf for Alcian Blue staining. (A-F) Representative images of Alcian Blue-stained pectoral fins at 4 dpf. (A) Uninjected WT/heterozygous (n=18). (B) Uninjected prdm1a−/− mutant (n=9). (C-F) prdm1a−/− mutants were injected with (C) WT hPRDM1 (n=10), (D) hPRDM1(p.C239Lfs*32) (n=10), (E) hPRDM1(p.T524R) (n=8) or (F) hPRDM1(p.T819A) mRNA (n=7). The representative uninjected control images in A and B are also used in Fig. S3, which shows the effect of overexpression of hPRDM1 variants in the WT background as part of the same experiment. Scale bars: 50 µm. (G-I) Measurements for the lengths of the (G) cleithrum, (H) endoskeletal disk and (I) fin fold and (J) the area of the scapulocoracoid and postcoracoid were averaged and compared using a one-way ANOVA, followed by a Tukey post-hoc test relative to uninjected prdm1a−/− mutants. Each dot represents one independent biological replicate. P-values are shown in the figure. Injection of WT hPRDM1 partially rescued the cleithrum, endoskeletal disk and fin fold of prdm1a−/− mutants. However, overexpression of the three SHFM variants failed to rescue the pectoral fin. Error bars represent the mean±s.d. cl, cleithrum; D, distal; ed, endoskeletal disk; ff, fin fold; Het, heterozygous; hpf, hours post fertilization; MT, prdm1a−/− mutant; P, proximal; pc, postcoracoid; sc, scapulocoracoid; WT, wildtype. |
|
Overexpression of Prdm1a using a global heat-shock Gal4/UAS system shows that proline/serine-rich and zinc finger domains are required for pectoral fin function. (A) Schematic of 4XnrUAS-modified prdm1a-2a-EGFP constructs that were injected into Tg(hsp70l:gal4FF);prdm1a+/− intercrosses. Results for the ability to rescue the pectoral fin are shown. (B) Experimental design for heat-shock Gal4/UAS rescue experiments. Following injection with the UAS construct, embryos at 6 hpf (shield stage) were heat shocked, leading to activation of Gal4, expression of the 4XnrUAS-modified prdm1a-2a-EGFP construct, and cleavage of the 2a viral peptide from EGFP. Embryos were screened for mosaic EGFP expression at 24 hpf. (C,D) Representative images of 24 hpf embryos injected with the 4XnrUAS-modified prdm1a-2a-EGFP construct at the single-cell stage. (C) No heat shock (control). (D) Mosaic EGFP expression in embryos that were injected and heat shocked. The dotted box marks the pectoral fin. Scale bars: 200 µm. (E-L) Representative images of Alcian Blue-stained pectoral fins at 4 dpf are shown. (E) Uninjected WT (n=36). (F) Uninjected prdm1a−/− mutants (n=11). (G-L) Mutants were injected with constructs containing (G) full-length Prdm1a (n=9), (H) Prdm1aΔSET (n=7), (I) Prdm1aΔP/S (n=13), (J) Prdm1aΔZnF (n=7), (K) Prdm1aΔP/S&ZnF (n=13) and (L) an EGFP negative control (n=16). Scale bars: 50 µm. The representative uninjected control images in E and F are also used in Fig. S4, which shows the effect of overexpression of modified Prdm1a in the WT background as part of the same experiment. (M-P) Measurements were taken for the length of the (M) cleithrum, (N) endoskeletal disk and (O) fin fold and (P) the area of the scapulocoracoid and postcoracoid. Each dot represents one independent biological replicate. Measurements for each individual were averaged and compared using a one-way ANOVA, followed by a Tukey's post-hoc test relative to uninjected, heat-shocked prdm1a−/− mutants. prdm1a−/− mutants injected with full-length Prdm1a exhibited a rescue in the endoskeletal disk (P=0.0654). Prdm1aΔSET also partially rescued the area of the scapulocoracoid/postcoracoid (P=0.0446). However, Prdm1aΔP/S, Prdm1aΔZnF and Prdm1aΔP/S&ZnF failed to rescue prdm1a−/− mutants. Error bars represent the mean±s.d. Δ, deleted; Ant, anterior; cl, cleithrum; D, distal; dpf, days post fertilization; e, eye; ed, endoskeletal disk; ff, fin fold; hpf, hours post fertilization; MT, prdm1a−/− mutant; P, proximal; pc, postcoracoid; pf, pectoral fin; Post, posterior; sc, scapulocoracoid; WT, wildtype; y, yolk. |
|
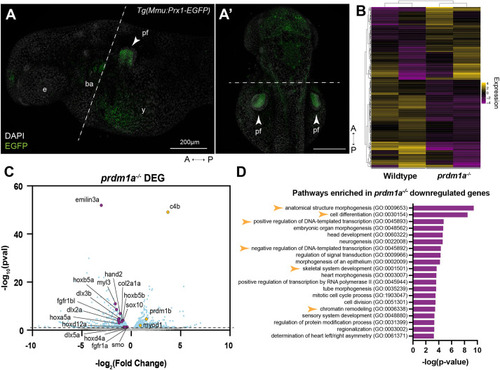
Loss of Prdm1a leads to downregulation of important limb development genes in the pectoral fin. RNA-seq was performed on isolated pectoral fin cells from about 250 WT and prdm1a−/− embryos at 48 hpf. (A) Lateral and (A′) dorsal view of EGFP-positive pectoral fins from the Tg(Mmu:Prx1-EGFP) zebrafish line at 48 hpf before FACS. Dashed lines indicate where the embryos were dissected prior to FACS. Scale bars: 200 µm. (B) Heat map of top 250 differentially expressed genes (Padj) between WT and prdm1a−/− embryos. (C) Volcano plot showing spread of differentially expressed genes in pectoral fins of prdm1a−/− compared to WT embryos. Light blue dots are significant, differentially expressed genes [−log10(P-value)≥1.15]. Purple dots are downregulated, selected genes of interest, whereas yellow dots are upregulated genes. (D) Downregulated genes in prdm1a−/− embryos were subjected to GO (Panther) pathway enrichment analysis. Yellow arrowheads highlight pathways of interest. A, anterior; ba, branchial arches; DEG, differentially expressed genes; e, eye; P, posterior; pf, pectoral fin; y, yolk. EXPRESSION / LABELING:
|
|
Prdm1a acts downstream of fin initiation and regulates Fgf signaling in the fin mesenchyme required for outgrowth and anterior/posterior patterning. (A,C,H,M,Q) Lateral views of pectoral fins from whole-mount WT and prdm1a−/− mutant embryos after hybridization chain reaction (HCR) was performed at 48 hpf. Scale bars: 50 µm. (A) tbx5a (pectoral fin initiation) and prdm1a expression (n=8 WT and 6 prdm1a−/− embryos). (B) Quantification of tbx5a expression using corrected total cell fluorescence (CTCF) showed no significant difference. (C) fgf10a (pectoral fin induction) and prdm1a expression (n=6 for each genotype). (D,E) Quantification of (D) fgf10a and (E) prdm1a expression along a line drawn from the most proximal to the most distal point of the fin bud using the line scan tool on ImageJ. Intensity and distance were normalized between 0 and 1. (F) Maximum normalized intensity of fgf10a shows a decrease in prdm1a−/− compared to WT embryos. (G) Length of the fin as measured by fgf10a gene expression. (H) fgf8a (AF outgrowth marker) and prdm1a expression (n=6 for each genotype). (I,J) Quantification of (I) fgf8a and (J) prdm1a expression along a line drawn from the most proximal to the most distal point of the fin bud. (K) Maximum normalized intensity of fgf8a shows a decrease in prdm1a−/− compared to WT embryos. (L) Length of the fin as measured by fgf8a gene expression. (M) dlx5a (outgrowth marker) and prdm1a expression (n=3 WT and 4 prdm1a−/− embryos). (N,O) Quantification of (N) dlx5a and (O) prdm1a expression along a line drawn from the most proximal to the most distal point of the fin bud. (P) Length of the cleithrum is decreased in prdm1a−/− compared to WT embryos (P=0.0143). (Q) shha (anterior/posterior patterning) and prdm1a expression (n=8 for each genotype). (R) Expression of shha was quantified using CTCF shows a decrease in prdm1a−/− compared to WT embryos (P=0.0294). Solid lines in line intensity graphs represent the mean±s.d. Statistical comparisons were made using unpaired, two-tailed, independent Student's t-test. All images are maximum projections of lateral views of the pectoral fin. The background was subtracted using the rolling ball feature in ImageJ (50 pixels). AF, apical fold; cl, cleithrum; CTCF, corrected total cell fluorescence; D, distal; hpf, hours post fertilization; P, proximal; ZPA, zone of polarizing activity. |
|
Prdm1a directly binds to and regulates limb genes. (A) Lateral view of EGFP-positive pectoral fins from the Tg(Mmu:Prx1-EGFP) zebrafish line at 24 hpf before CUT&RUN was performed. Scale bar: 200 µm. (B,C) Coverage heatmaps of (B) H3K27Ac and (C) Prdm1a binding across the genome 1.5 kb upstream and downstream of the peak center. (D) Annotation of enriched binding sites by Prdm1a. (E) Enriched Prdm1a peaks were subjected to Gene Ontology (GO) terms analysis using ChIPseeker's enrichGO function. (F) Prdm1a peaks were subjected to motif analysis using HOMER. The top ten motifs as well as known limb-related motifs are shown along with q-values. (G) Tracks showing H3K27Ac enrichment (open chromatin) and Prdm1a-binding sites for fgfr1a, dlx5a, dlx6a and smo. There is variability between replicates, but the overall trends are comparable (see Fig. S5). e, eye; pf, pectoral fin; TF, transcription factor; y, yolk. |
|
Working model of the Prdm1a gene regulatory network during pectoral fin development. Prdm1a acts downstream of pectoral fin initiation and tbx5a (Fig. 6A,B), but upstream of induction and Fgf signaling. Prdm1a directly binds to regulatory sequences of and activates fgfr1a (Fig. 7G), allowing Fgf10a to bind (Fig. 6C-G). Fgf10a then activates Fgf8a in the apical fold (Fig. 6H-L), signaling pectoral fin outgrowth and differentiation. Prdm1a also directly binds to putative enhancers and promoter regions of smo (Fig. 7G), a receptor in Shh signaling. shha is expressed in the zone of polarizing activity (Fig. 6Q,R). It is required for anterior/posterior patterning as well as regulating fgf8a expression. Dashed arrows illustrate additional feedback loops that have been demonstrated in mice and/or chick but have not yet been shown in zebrafish. Finally, Prdm1a directly binds to putative enhancers of dlx5a and dlx6a, additional fin outgrowth markers (Fig. 6M-P). |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |